Accurate and Fast Deep Learning Dose Prediction for a Preclinical Microbeam Radiation Therapy Study Using Low-Statistics Monte Carlo Simulations
- PMID: 37046798
- PMCID: PMC10093595
- DOI: 10.3390/cancers15072137
Accurate and Fast Deep Learning Dose Prediction for a Preclinical Microbeam Radiation Therapy Study Using Low-Statistics Monte Carlo Simulations
Abstract
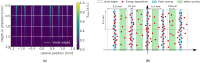
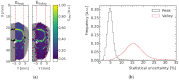
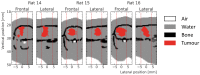
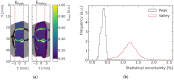
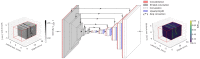
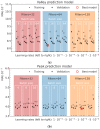
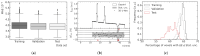
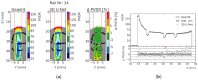
Microbeam radiation therapy (MRT) utilizes coplanar synchrotron radiation beamlets and is a proposed treatment approach for several tumor diagnoses that currently have poor clinical treatment outcomes, such as gliosarcomas. Monte Carlo (MC) simulations are one of the most used methods at the Imaging and Medical Beamline, Australian Synchrotron to calculate the dose in MRT preclinical studies. The steep dose gradients associated with the 50μm-wide coplanar beamlets present a significant challenge for precise MC simulation of the dose deposition of an MRT irradiation treatment field in a short time frame. The long computation times inhibit the ability to perform dose optimization in treatment planning or apply online image-adaptive radiotherapy techniques to MRT. Much research has been conducted on fast dose estimation methods for clinically available treatments. However, such methods, including GPU Monte Carlo implementations and machine learning (ML) models, are unavailable for novel and emerging cancer radiotherapy options such as MRT. In this work, the successful application of a fast and accurate ML dose prediction model for a preclinical MRT rodent study is presented for the first time. The ML model predicts the peak doses in the path of the microbeams and the valley doses between them, delivered to the tumor target in rat patients. A CT imaging dataset is used to generate digital phantoms for each patient. Augmented variations of the digital phantoms are used to simulate with Geant4 the energy depositions of an MRT beam inside the phantoms with 15% (high-noise) and 2% (low-noise) statistical uncertainty. The high-noise MC simulation data are used to train the ML model to predict the energy depositions in the digital phantoms. The low-noise MC simulations data are used to test the predictive power of the ML model. The predictions of the ML model show an agreement within 3% with low-noise MC simulations for at least 77.6% of all predicted voxels (at least 95.9% of voxels containing tumor) in the case of the valley dose prediction and for at least 93.9% of all predicted voxels (100.0% of voxels containing tumor) in the case of the peak dose prediction. The successful use of high-noise MC simulations for the training, which are much faster to produce, accelerates the production of the training data of the ML model and encourages transfer of the ML model to different treatment modalities for other future applications in novel radiation cancer therapies.
Keywords: Geant4; Monte Carlo simulation; deep learning; dose prediction; microbeam radiation therapy; preclinical study.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Fast and accurate dose predictions for novel radiotherapy treatments in heterogeneous phantoms using conditional 3D-UNet generative adversarial networks.Med Phys. 2022 May;49(5):3389-3404. doi: 10.1002/mp.15555. Epub 2022 Mar 3. Med Phys. 2022. PMID: 35184310
-
Monte Carlo-based treatment planning system calculation engine for microbeam radiation therapy.Med Phys. 2012 May;39(5):2829-38. doi: 10.1118/1.4705351. Med Phys. 2012. PMID: 22559655
-
A high-resolution dose calculation engine for X-ray microbeams radiation therapy.Med Phys. 2022 Jun;49(6):3999-4017. doi: 10.1002/mp.15637. Epub 2022 Apr 12. Med Phys. 2022. PMID: 35342953 Free PMC article.
-
Technical advances in x-ray microbeam radiation therapy.Phys Med Biol. 2020 Jan 17;65(2):02TR01. doi: 10.1088/1361-6560/ab5507. Phys Med Biol. 2020. PMID: 31694009 Review.
-
Effects of pulsed, spatially fractionated, microscopic synchrotron X-ray beams on normal and tumoral brain tissue.Mutat Res. 2010 Apr-Jun;704(1-3):160-6. doi: 10.1016/j.mrrev.2009.12.003. Epub 2009 Dec 23. Mutat Res. 2010. PMID: 20034592 Review.
References
-
- Pastor-Serrano O., Perkó Z. Learning the Physics of Particle Transport via Transformers. arXiv. 2021 doi: 10.1609/aaai.v36i11.21466.2109.03951 - DOI
LinkOut - more resources
Full Text Sources

