Lipid Metabolic Reprogramming in Embryonal Neoplasms with MYCN Amplification
- PMID: 37046804
- PMCID: PMC10093342
- DOI: 10.3390/cancers15072144
Lipid Metabolic Reprogramming in Embryonal Neoplasms with MYCN Amplification
Abstract
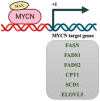
Tumor cells reprogram their metabolism, including glucose, glutamine, nucleotide, lipid, and amino acids to meet their enhanced energy demands, redox balance, and requirement of biosynthetic substrates for uncontrolled cell proliferation. Altered lipid metabolism in cancer provides lipids for rapid membrane biogenesis, generates the energy required for unrestricted cell proliferation, and some of the lipids act as signaling pathway mediators. In this review, we focus on the role of lipid metabolism in embryonal neoplasms with MYCN dysregulation. We specifically review lipid metabolic reactions in neuroblastoma, retinoblastoma, medulloblastoma, Wilms tumor, and rhabdomyosarcoma and the possibility of targeting lipid metabolism. Additionally, the regulation of lipid metabolism by the MYCN oncogene is discussed.
Keywords: MYCN; cancer; embryonal tumors; lipid metabolism; therapeutic targeting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The MYCN Protein in Health and Disease.Genes (Basel). 2017 Mar 30;8(4):113. doi: 10.3390/genes8040113. Genes (Basel). 2017. PMID: 28358317 Free PMC article. Review.
-
MYCN and Metabolic Reprogramming in Neuroblastoma.Cancers (Basel). 2022 Aug 25;14(17):4113. doi: 10.3390/cancers14174113. Cancers (Basel). 2022. PMID: 36077650 Free PMC article. Review.
-
MYCN expression in human rhabdomyosarcoma cell lines and tumour samples.J Pathol. 2002 Apr;196(4):450-8. doi: 10.1002/path.1068. J Pathol. 2002. PMID: 11920742
-
Childhood tumors of the nervous system as disorders of normal development.Curr Opin Pediatr. 2006 Dec;18(6):634-8. doi: 10.1097/MOP.0b013e32801080fe. Curr Opin Pediatr. 2006. PMID: 17099362 Review.
-
Metabolic profiling of the three neural derived embryonal pediatric tumors retinoblastoma, neuroblastoma and medulloblastoma, identifies distinct metabolic profiles.Oncotarget. 2018 Jan 11;9(13):11336-11351. doi: 10.18632/oncotarget.24168. eCollection 2018 Feb 16. Oncotarget. 2018. PMID: 29541417 Free PMC article.
Cited by
-
Evolving Diagnostic and Treatment Strategies for Pediatric CNS Tumors: The Impact of Lipid Metabolism.Biomedicines. 2023 May 5;11(5):1365. doi: 10.3390/biomedicines11051365. Biomedicines. 2023. PMID: 37239036 Free PMC article. Review.
-
Molecular pathway of anticancer effect of next-generation HSP90 inhibitors XL-888 and Debio0932 in neuroblastoma cell line.Med Oncol. 2024 Jul 3;41(8):194. doi: 10.1007/s12032-024-02428-z. Med Oncol. 2024. PMID: 38958814 Free PMC article.
-
Targeting tumor angiogenesis and metabolism: a new perspective in pediatric thoracic tumor therapy.Front Cell Dev Biol. 2025 Mar 27;13:1558403. doi: 10.3389/fcell.2025.1558403. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40213391 Free PMC article. Review.
-
Deciphering Medulloblastoma: Epigenetic and Metabolic Changes Driving Tumorigenesis and Treatment Outcomes.Biomedicines. 2025 Aug 4;13(8):1898. doi: 10.3390/biomedicines13081898. Biomedicines. 2025. PMID: 40868156 Free PMC article. Review.
References
-
- Schwab M., Alitalo K., Klempnauer K.H., Varmus H.E., Bishop J.M., Gilbert F., Brodeur G., Goldstein M., Trent J. Amplified DNA with Limited Homology to Myc Cellular Oncogene Is Shared by Human Neuroblastoma Cell Lines and a Neuroblastoma Tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

