Evodiamine Exhibits Anti-Bladder Cancer Activity by Suppression of Glutathione Peroxidase 4 and Induction of Ferroptosis
- PMID: 37046995
- PMCID: PMC10094601
- DOI: 10.3390/ijms24076021
Evodiamine Exhibits Anti-Bladder Cancer Activity by Suppression of Glutathione Peroxidase 4 and Induction of Ferroptosis
Abstract
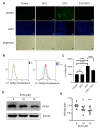
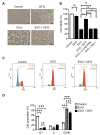
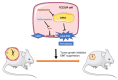
Evodiamine (EVO) exhibits anti-cancer activity through the inhibition of cell proliferation; however, little is known about its underlying mechanism. To determine whether ferroptosis is involved in the therapeutic effects of EVO, we investigated critical factors, such as lipid peroxidation levels and glutathione peroxidase 4 (GPX4) expression, under EVO treatment. Our results showed that EVO inhibited the cell proliferation of poorly differentiated, high-grade bladder cancer TCCSUP cells in a dose- and time-dependent manner. Lipid peroxides were detected by fluorescence microscopy after cancer cell exposure to EVO. GPX4, which catalyzes the conversion of lipid peroxides to prevent cells from undergoing ferroptosis, was decreased dose-dependently by EVO treatment. Given the features of iron dependency and lipid-peroxidation-driven death in ferroptosis, the iron chelator deferoxamine (DFO) was used to suppress EVO-induced ferroptosis. The lipid peroxide level significantly decreased when cells were treated with DFO prior to EVO treatment. DFO also attenuated EVO-induced cell death. Co-treatment with a pan-caspase inhibitor or necroptosis inhibitor with EVO did not alleviate cancer cell death. These results indicate that EVO induces ferroptosis rather than apoptosis or necroptosis. Furthermore, EVO suppressed the migratory ability, decreased the expression of mesenchymal markers, and increased epithelial marker expression, determined by a transwell migration assay and Western blotting. The TCCSUP bladder tumor xenograft tumor model confirmed the effects of EVO on the inhibition of tumor growth and EMT. In conclusion, EVO is a novel inducer for activating the ferroptosis of bladder cancer cells and may be a potential therapeutic agent for bladder cancer.
Keywords: bladder cancer; evodiamine; ferroptosis; glutathione peroxidase 4; lipid peroxidation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Significance of glutathione peroxidase 4 and intracellular iron level in ovarian cancer cells-"utilization" of ferroptosis mechanism.Inflamm Res. 2021 Dec;70(10-12):1177-1189. doi: 10.1007/s00011-021-01495-6. Epub 2021 Sep 19. Inflamm Res. 2021. PMID: 34537856
-
The glutathione peroxidase Gpx4 prevents lipid peroxidation and ferroptosis to sustain Treg cell activation and suppression of antitumor immunity.Cell Rep. 2021 Jun 15;35(11):109235. doi: 10.1016/j.celrep.2021.109235. Cell Rep. 2021. PMID: 34133924
-
Solasonine promotes ferroptosis of hepatoma carcinoma cells via glutathione peroxidase 4-induced destruction of the glutathione redox system.Biomed Pharmacother. 2020 Sep;129:110282. doi: 10.1016/j.biopha.2020.110282. Epub 2020 Jun 9. Biomed Pharmacother. 2020. PMID: 32531676
-
A white paper on Phospholipid Hydroperoxide Glutathione Peroxidase (GPx4) forty years later.Free Radic Biol Med. 2022 Aug 1;188:117-133. doi: 10.1016/j.freeradbiomed.2022.06.227. Epub 2022 Jun 16. Free Radic Biol Med. 2022. PMID: 35718302 Review.
-
Prospects for Anti-Tumor Mechanism and Potential Clinical Application Based on Glutathione Peroxidase 4 Mediated Ferroptosis.Int J Mol Sci. 2023 Jan 13;24(2):1607. doi: 10.3390/ijms24021607. Int J Mol Sci. 2023. PMID: 36675129 Free PMC article. Review.
Cited by
-
Evodiamine induces ferroptosis in prostate cancer cells by inhibiting TRIM26-mediated stabilization of GPX4.Chin Med. 2025 May 26;20(1):71. doi: 10.1186/s13020-025-01130-0. Chin Med. 2025. PMID: 40420092 Free PMC article.
-
Evodiamine Boosts AR Expression to Trigger Senescence and Halt Proliferation in OSCC Cells.Curr Issues Mol Biol. 2025 Jul 17;47(7):558. doi: 10.3390/cimb47070558. Curr Issues Mol Biol. 2025. PMID: 40729027 Free PMC article.
-
Molecular mechanisms of ferroptosis and its effects on bladder cancer.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 Feb 28;49(2):286-295. doi: 10.11817/j.issn.1672-7347.2024.230352. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38755725 Free PMC article. Review. Chinese, English.
-
Ferroptosis: An Emerging Target for Bladder Cancer Therapy.Curr Issues Mol Biol. 2023 Oct 10;45(10):8201-8214. doi: 10.3390/cimb45100517. Curr Issues Mol Biol. 2023. PMID: 37886960 Free PMC article. Review.
-
The mechanism of ferroptosis and its related diseases.Mol Biomed. 2023 Oct 16;4(1):33. doi: 10.1186/s43556-023-00142-2. Mol Biomed. 2023. PMID: 37840106 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

