Hydroperoxidation of Docosahexaenoic Acid by Human ALOX12 and pigALOX15-mini-LOX
- PMID: 37047037
- PMCID: PMC10094721
- DOI: 10.3390/ijms24076064
Hydroperoxidation of Docosahexaenoic Acid by Human ALOX12 and pigALOX15-mini-LOX
Abstract
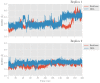
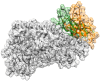
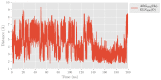
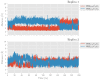
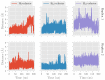
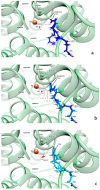
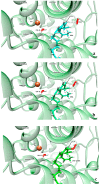
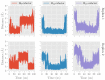
Human lipoxygenase 12 (hALOX12) catalyzes the conversion of docosahexaenoic acid (DHA) into mainly 14S-hydroperoxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14S-H(p)DHA). This hydroperoxidation reaction is followed by an epoxidation and hydrolysis process that finally leads to maresin 1 (MaR1), a potent bioactive specialized pro-resolving mediator (SPM) in chronic inflammation resolution. By combining docking, molecular dynamics simulations, and quantum mechanics/molecular mechanics calculations, we have computed the potential energy profile of DHA hydroperoxidation in the active site of hALOX12. Our results describe the structural evolution of the molecular system at each step of this catalytic reaction pathway. Noteworthy, the required stereospecificity of the reaction leading to MaR1 is explained by the configurations adopted by DHA bound to hALOX12, along with the stereochemistry of the pentadienyl radical formed after the first step of the mechanism. In pig lipoxygenase 15 (pigALOX15-mini-LOX), our calculations suggest that 14S-H(p)DHA can be formed, but with a stereochemistry that is inadequate for MaR1 biosynthesis.
Keywords: QM/MM calculations; enzyme catalysis; human platelet ALOX12; hydroperoxidation mechanism; molecular dynamics simulations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













Similar articles
-
Biosynthesis of the Maresin Intermediate, 13S,14S-Epoxy-DHA, by Human 15-Lipoxygenase and 12-Lipoxygenase and Its Regulation through Negative Allosteric Modulators.Biochemistry. 2020 May 19;59(19):1832-1844. doi: 10.1021/acs.biochem.0c00233. Epub 2020 May 7. Biochemistry. 2020. PMID: 32324389 Free PMC article.
-
15-Lipoxygenase-1 biosynthesis of 7S,14S-diHDHA implicates 15-lipoxygenase-2 in biosynthesis of resolvin D5.J Lipid Res. 2020 Jul;61(7):1087-1103. doi: 10.1194/jlr.RA120000777. Epub 2020 May 13. J Lipid Res. 2020. PMID: 32404334 Free PMC article.
-
The novel 13S,14S-epoxy-maresin is converted by human macrophages to maresin 1 (MaR1), inhibits leukotriene A4 hydrolase (LTA4H), and shifts macrophage phenotype.FASEB J. 2013 Jul;27(7):2573-83. doi: 10.1096/fj.13-227728. Epub 2013 Mar 15. FASEB J. 2013. PMID: 23504711 Free PMC article.
-
DHA metabolism: targeting the brain and lipoxygenation.Mol Neurobiol. 2010 Aug;42(1):48-51. doi: 10.1007/s12035-010-8131-7. Epub 2010 Apr 28. Mol Neurobiol. 2010. PMID: 20422316 Free PMC article. Review.
-
The eosinophil chemoattractant 5-oxo-ETE and the OXE receptor.Prog Lipid Res. 2013 Oct;52(4):651-65. doi: 10.1016/j.plipres.2013.09.001. Epub 2013 Sep 19. Prog Lipid Res. 2013. PMID: 24056189 Free PMC article. Review.
Cited by
-
Construction of a Novel Mitochondria-Associated Gene Model for Assessing ESCC Immune Microenvironment and Predicting Survival.J Microbiol Biotechnol. 2024 May 28;34(5):1164-1177. doi: 10.4014/jmb.2310.10052. Epub 2024 Feb 22. J Microbiol Biotechnol. 2024. PMID: 38719775 Free PMC article.
References
-
- Steinhilber D. Lipoxygenases: An Introduction. In: Steinhilber D., editor. Lipoxygenases in Inflammation. Springer; Cham, Switzerland: 2016. pp. 1–6.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

