Accurate Prediction of Transcriptional Activity of Single Missense Variants in HIV Tat with Deep Learning
- PMID: 37047108
- PMCID: PMC10093788
- DOI: 10.3390/ijms24076138
Accurate Prediction of Transcriptional Activity of Single Missense Variants in HIV Tat with Deep Learning
Abstract
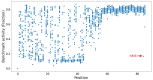
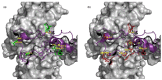
Tat is an essential gene for increasing the transcription of all HIV genes, and affects HIV replication, HIV exit from latency, and AIDS progression. The Tat gene frequently mutates in vivo and produces variants with diverse activities, contributing to HIV viral heterogeneity as well as drug-resistant clones. Thus, identifying the transcriptional activities of Tat variants will help to better understand AIDS pathology and treatment. We recently reported the missense mutation landscape of all single amino acid Tat variants. In these experiments, a fraction of double missense alleles exhibited intragenic epistasis. However, it is too time-consuming and costly to determine the effect of the variants for all double mutant alleles through experiments. Therefore, we propose a combined GigaAssay/deep learning approach. As a first step to determine activity landscapes for complex variants, we evaluated a deep learning framework using previously reported GigaAssay experiments to predict how transcription activity is affected by Tat variants with single missense substitutions. Our approach achieved a 0.94 Pearson correlation coefficient when comparing the predicted to experimental activities. This hybrid approach can be extensible to more complex Tat alleles for a better understanding of the genetic control of HIV genome transcription.
Keywords: HIV; deep learning; tat protein; variant effect.
Conflict of interest statement
Martin R. Schiller and CJ Giacoletto are associated with Heligenics, a company pursuing commercial interests for the GigaAssay.
Figures



Similar articles
-
GigaAssay - An adaptable high-throughput saturation mutagenesis assay platform.Genomics. 2022 Jul;114(4):110439. doi: 10.1016/j.ygeno.2022.110439. Epub 2022 Jul 26. Genomics. 2022. PMID: 35905834 Free PMC article.
-
Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia.J Neurovirol. 2011 Feb;17(1):70-81. doi: 10.1007/s13365-010-0002-5. Epub 2010 Nov 30. J Neurovirol. 2011. PMID: 21165788
-
HIV latency reversing agents act through Tat post translational modifications.Retrovirology. 2018 May 11;15(1):36. doi: 10.1186/s12977-018-0421-6. Retrovirology. 2018. PMID: 29751762 Free PMC article.
-
Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription.Curr Pharm Des. 2017;23(28):4079-4090. doi: 10.2174/1381612823666170622104355. Curr Pharm Des. 2017. PMID: 28641539 Free PMC article. Review.
-
Genetic variation and function of the HIV-1 Tat protein.Med Microbiol Immunol. 2019 Apr;208(2):131-169. doi: 10.1007/s00430-019-00583-z. Epub 2019 Mar 5. Med Microbiol Immunol. 2019. PMID: 30834965 Free PMC article. Review.
Cited by
-
Accurate prediction of functional effect of single amino acid variants with deep learning.Comput Struct Biotechnol J. 2023 Nov 10;21:5776-5784. doi: 10.1016/j.csbj.2023.11.017. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38074467 Free PMC article.
-
MMRT: MultiMut Recursive Tree for predicting functional effects of high-order protein variants from low-order variants.Comput Struct Biotechnol J. 2025 Feb 18;27:672-681. doi: 10.1016/j.csbj.2025.02.012. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40070521 Free PMC article.
References
-
- Basic Statistics|HIV Basics|HIV/AIDS|CDC. [(accessed on 6 May 2022)]; Available online: https://www.cdc.gov/hiv/basics/statistics.html.
-
- Palmer S., Kearney M., Maldarelli F., Halvas E.K., Bixby C.J., Bazmi H., Rock D., Falloon J., Davey R.T., Jr., Dewar R.L., et al. Multiple, Linked Human Immunodeficiency Virus Type 1 Drug Resistance Mutations in Treatment-Experienced Patients Are Missed by Standard Genotype Analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

