Allosteric Regulation of G-Protein-Coupled Receptors: From Diversity of Molecular Mechanisms to Multiple Allosteric Sites and Their Ligands
- PMID: 37047169
- PMCID: PMC10094638
- DOI: 10.3390/ijms24076187
Allosteric Regulation of G-Protein-Coupled Receptors: From Diversity of Molecular Mechanisms to Multiple Allosteric Sites and Their Ligands
Abstract
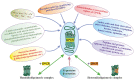
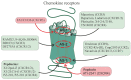
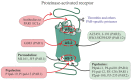
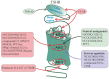
Allosteric regulation is critical for the functioning of G protein-coupled receptors (GPCRs) and their signaling pathways. Endogenous allosteric regulators of GPCRs are simple ions, various biomolecules, and protein components of GPCR signaling (G proteins and β-arrestins). The stability and functional activity of GPCR complexes is also due to multicenter allosteric interactions between protomers. The complexity of allosteric effects caused by numerous regulators differing in structure, availability, and mechanisms of action predetermines the multiplicity and different topology of allosteric sites in GPCRs. These sites can be localized in extracellular loops; inside the transmembrane tunnel and in its upper and lower vestibules; in cytoplasmic loops; and on the outer, membrane-contacting surface of the transmembrane domain. They are involved in the regulation of basal and orthosteric agonist-stimulated receptor activity, biased agonism, GPCR-complex formation, and endocytosis. They are targets for a large number of synthetic allosteric regulators and modulators, including those constructed using molecular docking. The review is devoted to the principles and mechanisms of GPCRs allosteric regulation, the multiplicity of allosteric sites and their topology, and the endogenous and synthetic allosteric regulators, including autoantibodies and pepducins. The allosteric regulation of chemokine receptors, proteinase-activated receptors, thyroid-stimulating and luteinizing hormone receptors, and beta-adrenergic receptors are described in more detail.
Keywords: G protein-coupled receptor; allosteric modulator; allosteric site; autoantibody; chemokine receptor; heterotrimeric G protein; luteinizing hormone receptor; pepducin; proteinase-activated receptor; thyroid-stimulating hormone receptor.
Conflict of interest statement
The author declares no conflict of interest.
Figures
Similar articles
-
Designing Structure-specific and Switchable Allosteric Effectors for GPCRs Based on the Causality and Energetics of Inherent Signaling.J Mol Biol. 2025 Jun 11:169293. doi: 10.1016/j.jmb.2025.169293. Online ahead of print. J Mol Biol. 2025. PMID: 40513649
-
The impact of cryo-EM on determining allosteric modulator-bound structures of G protein-coupled receptors.Curr Opin Struct Biol. 2023 Apr;79:102560. doi: 10.1016/j.sbi.2023.102560. Epub 2023 Feb 26. Curr Opin Struct Biol. 2023. PMID: 36848776 Review.
-
Deciphering complexity of GPCR signaling and modulation: implications and perspectives for drug discovery.Clin Sci (Lond). 2025 May 20;139(10):463-77. doi: 10.1042/CS20245182. Clin Sci (Lond). 2025. PMID: 40400289 Free PMC article. Review.
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery.Signal Transduct Target Ther. 2024 Apr 10;9(1):88. doi: 10.1038/s41392-024-01803-6. Signal Transduct Target Ther. 2024. PMID: 38594257 Free PMC article. Review.
-
Large scale investigation of GPCR molecular dynamics data uncovers allosteric sites and lateral gateways.Nat Commun. 2025 Feb 27;16(1):2020. doi: 10.1038/s41467-025-57034-y. Nat Commun. 2025. PMID: 40016203 Free PMC article.
Cited by
-
An orthosteric/allosteric bivalent peptide agonist comprising covalently linked protease-activated receptor-derived peptides mimics in vitro and in vivo activities of activated protein C.J Thromb Haemost. 2024 Jul;22(7):2039-2051. doi: 10.1016/j.jtha.2024.04.007. Epub 2024 Apr 24. J Thromb Haemost. 2024. PMID: 38670314 Free PMC article.
-
Structural and dynamic insights into the activation of the μ-opioid receptor by an allosteric modulator.Nat Commun. 2024 May 13;15(1):3544. doi: 10.1038/s41467-024-47792-6. Nat Commun. 2024. PMID: 38740791 Free PMC article.
-
Charge Movements and Conformational Changes: Biophysical Properties and Physiology of Voltage-Dependent GPCRs.Biomolecules. 2024 Dec 23;14(12):1652. doi: 10.3390/biom14121652. Biomolecules. 2024. PMID: 39766359 Free PMC article. Review.
-
Effect of a Low-Molecular-Weight Allosteric Agonist of the Thyroid-Stimulating Hormone Receptor on Basal and Thyroliberin-Stimulated Activity of Thyroid System in Diabetic Rats.Int J Mol Sci. 2025 Jan 15;26(2):703. doi: 10.3390/ijms26020703. Int J Mol Sci. 2025. PMID: 39859419 Free PMC article.
-
Structural perspectives on chemokine receptors.Biochem Soc Trans. 2024 Jun 26;52(3):1011-1024. doi: 10.1042/BST20230358. Biochem Soc Trans. 2024. PMID: 38856028 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

