Profiling and Cellular Analyses of Obesity-Related circRNAs in Neurons and Glia under Obesity-like In Vitro Conditions
- PMID: 37047207
- PMCID: PMC10094513
- DOI: 10.3390/ijms24076235
Profiling and Cellular Analyses of Obesity-Related circRNAs in Neurons and Glia under Obesity-like In Vitro Conditions
Abstract
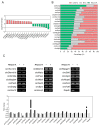
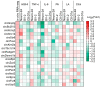
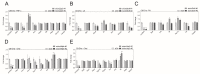
Recent evidence indicates that the pathogenesis of neurodegenerative diseases, including Alzheimer's disease, is associated with metabolic disorders such as diabetes and obesity. Various circular RNAs (circRNAs) have been found in brain tissues and recent studies have suggested that circRNAs are related to neuropathological mechanisms in the brain. However, there is a lack of interest in the involvement of circRNAs in metabolic imbalance-related neuropathological problems until now. Herein we profiled and analyzed diverse circRNAs in mouse brain cell lines (Neuro-2A neurons, BV-2 microglia, and C8-D1a astrocytes) exposed to obesity-related in vitro conditions (high glucose, high insulin, and high levels of tumor necrosis factor-alpha, interleukin 6, palmitic acid, linoleic acid, and cholesterol). We observed that various circRNAs were differentially expressed according to cell types with many of these circRNAs conserved in humans. After suppressing the expression of these circRNAs using siRNAs, we observed that these circRNAs regulate genes related to inflammatory responses, formation of synaptic vesicles, synaptic density, and fatty acid oxidation in neurons; scavenger receptors in microglia; and fatty acid signaling, inflammatory signaling cyto that may play important roles in metabolic disorders associated with neurodegenerative diseases.
Keywords: astrocytes; circular RNAs (circRNAs); microglia; neurons; obesity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Exploring the Regulatory Role of Circular RNAs in Neurodegenerative Disorders.Int J Mol Sci. 2019 Nov 4;20(21):5477. doi: 10.3390/ijms20215477. Int J Mol Sci. 2019. PMID: 31689888 Free PMC article. Review.
-
Recent insights into the roles of circular RNAs in human brain development and neurologic diseases.Int J Biol Macromol. 2023 Jan 15;225:1038-1048. doi: 10.1016/j.ijbiomac.2022.11.166. Epub 2022 Nov 19. Int J Biol Macromol. 2023. PMID: 36410538 Review.
-
The Emerging Role and Promise of Circular RNAs in Obesity and Related Metabolic Disorders.Cells. 2020 Jun 16;9(6):1473. doi: 10.3390/cells9061473. Cells. 2020. PMID: 32560220 Free PMC article. Review.
-
Cell type-specific circular RNA expression in human glial cells.Genomics. 2020 Nov;112(6):5265-5274. doi: 10.1016/j.ygeno.2020.09.042. Epub 2020 Sep 20. Genomics. 2020. PMID: 32966858
-
Emergent Roles of Circular RNAs in Metabolism and Metabolic Disorders.Int J Mol Sci. 2022 Jan 18;23(3):1032. doi: 10.3390/ijms23031032. Int J Mol Sci. 2022. PMID: 35162956 Free PMC article. Review.
Cited by
-
Bioinformatics-led identification of pathophysiological hallmark genes in diabesotension via graph clustering method.J Diabetes Metab Disord. 2025 Jun 7;24(1):141. doi: 10.1007/s40200-025-01659-9. eCollection 2025 Jun. J Diabetes Metab Disord. 2025. PMID: 40491693
-
High-fat diet and chronic restraint stress exacerbate anxiety-depressive behaviors via astrocytic A1 phenotype transformation.Sci Rep. 2025 Apr 29;15(1):15031. doi: 10.1038/s41598-025-99355-4. Sci Rep. 2025. PMID: 40301496 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

