Ab Initio Modelling of the Structure of ToxA-like and MAX Fungal Effector Proteins
- PMID: 37047233
- PMCID: PMC10094246
- DOI: 10.3390/ijms24076262
Ab Initio Modelling of the Structure of ToxA-like and MAX Fungal Effector Proteins
Abstract
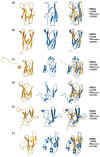
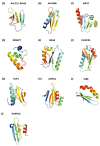
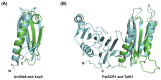
Pathogenic fungal diseases in crops are mediated by the release of effector proteins that facilitate infection. Characterising the structure of these fungal effectors is vital to understanding their virulence mechanisms and interactions with their hosts, which is crucial in the breeding of plant cultivars for disease resistance. Several effectors have been identified and validated experimentally; however, their lack of sequence conservation often impedes the identification and prediction of their structure using sequence similarity approaches. Structural similarity has, nonetheless, been observed within fungal effector protein families, creating interest in validating the use of computational methods to predict their tertiary structure from their sequence. We used Rosetta ab initio modelling to predict the structures of members of the ToxA-like and MAX effector families for which experimental structures are known to validate this method. An optimised approach was then used to predict the structures of phenotypically validated effectors lacking known structures. Rosetta was found to successfully predict the structure of fungal effectors in the ToxA-like and MAX families, as well as phenotypically validated but structurally unconfirmed effector sequences. Interestingly, potential new effector structural families were identified on the basis of comparisons with structural homologues and the identification of associated protein domains.
Keywords: MAX effector family; MaxCluster; Rosetta; TM-score; ToxA-like effector family; ab initio modelling; fungal effector protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Park C., Chen S., Shirsekar G., Zhou B., Khang C., Songkumarn P., Afzal A., Ning Y., Wang R., Bellizzi M., et al. The magnaporthe oryzae effector avrpiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

