Plasma Proteomics Unveil Novel Immune Signatures and Biomarkers upon SARS-CoV-2 Infection
- PMID: 37047248
- PMCID: PMC10093853
- DOI: 10.3390/ijms24076276
Plasma Proteomics Unveil Novel Immune Signatures and Biomarkers upon SARS-CoV-2 Infection
Abstract
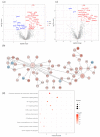
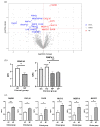
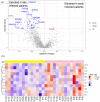
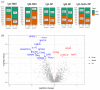
Several elements have an impact on COVID-19, including comorbidities, age and sex. To determine the protein profile changes in peripheral blood caused by a SARS-CoV-2 infection, a proximity extension assay was used to quantify 1387 proteins in plasma samples among 28 Finnish patients with COVID-19 with and without comorbidities and their controls. Key immune signatures, including CD4 and CD28, were changed in patients with comorbidities. Importantly, several unreported elevated proteins in patients with COVID-19, such as RBP2 and BST2, which show anti-microbial activity, along with proteins involved in extracellular matrix remodeling, including MATN2 and COL6A3, were identified. RNF41 was downregulated in patients compared to healthy controls. Our study demonstrates that SARS-CoV-2 infection causes distinct plasma protein changes in the presence of comorbidities despite the interpatient heterogeneity, and several novel potential biomarkers associated with a SARS-CoV-2 infection alone and in the presence of comorbidities were identified. Protein changes linked to the generation of SARS-CoV-2-specific antibodies, long-term effects and potential association with post-COVID-19 condition were revealed. Further study to characterize the identified plasma protein changes from larger cohorts with more diverse ethnicities of patients with COVID-19 combined with functional studies will facilitate the identification of novel diagnostic, prognostic biomarkers and potential therapeutic targets for patients with COVID-19.
Keywords: COVID-19; SARS-CoV-2; biomarker; immune signature; plasma proteomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers.Biomedicines. 2024 Apr 10;12(4):840. doi: 10.3390/biomedicines12040840. Biomedicines. 2024. PMID: 38672194 Free PMC article.
-
Plasma proteomics identify potential severity biomarkers from COVID-19 associated network.Proteomics Clin Appl. 2023 Mar;17(2):e2200070. doi: 10.1002/prca.202200070. Epub 2022 Oct 25. Proteomics Clin Appl. 2023. PMID: 36217943 Free PMC article.
-
Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection.Viruses. 2021 Dec 7;13(12):2456. doi: 10.3390/v13122456. Viruses. 2021. PMID: 34960725 Free PMC article.
-
Progress in understanding COVID-19: insights from the omics approach.Crit Rev Clin Lab Sci. 2021 Jun;58(4):242-252. doi: 10.1080/10408363.2020.1851167. Epub 2020 Dec 29. Crit Rev Clin Lab Sci. 2021. PMID: 33375876 Review.
-
Blood proteomics of COVID-19 infection: An update.Clin Chim Acta. 2024 Aug 15;562:119881. doi: 10.1016/j.cca.2024.119881. Epub 2024 Jul 19. Clin Chim Acta. 2024. PMID: 39033952 Review.
Cited by
-
Short Survey on the Protein Modifications in Plasma during SARS-CoV-2 Infection.Int J Mol Sci. 2023 Sep 14;24(18):14109. doi: 10.3390/ijms241814109. Int J Mol Sci. 2023. PMID: 37762413 Free PMC article.
-
A reduced proteomic signature in critically ill Covid-19 patients determined with plasma antibody micro-array and machine learning.Clin Proteomics. 2024 May 17;21(1):33. doi: 10.1186/s12014-024-09488-3. Clin Proteomics. 2024. PMID: 38760690 Free PMC article.
-
Coronavirus Disease Pathophysiology: Biomarkers, Potential New Remedies, Comorbidities, Long COVID-19, Post Pandemic Epidemiological Surveillance.Int J Mol Sci. 2023 Jul 31;24(15):12236. doi: 10.3390/ijms241512236. Int J Mol Sci. 2023. PMID: 37569612 Free PMC article.
-
Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers.Biomedicines. 2024 Apr 10;12(4):840. doi: 10.3390/biomedicines12040840. Biomedicines. 2024. PMID: 38672194 Free PMC article.
-
Plasma proteomic profiling of hospitalized patients co-infected with HIV and SARS-CoV-2.Front Immunol. 2025 Jun 11;16:1601672. doi: 10.3389/fimmu.2025.1601672. eCollection 2025. Front Immunol. 2025. PMID: 40568587 Free PMC article.
References
-
- WHO . COVID-19 Weekly Epidemiological Update. World Health Organization; Geneva, Switzerland: 2022. pp. 1–23.
-
- Bergamaschi L., Mescia F., Turner L., Hanson A.L., Kotagiri P., Dunmore B.J., Ruffieux H., De Sa A., Huhn O., Morgan M.D., et al. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity. 2021;54:1257–1275.e8. doi: 10.1016/j.immuni.2021.05.010. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

