Next Generation of Ovarian Cancer Detection Using Aptamers
- PMID: 37047289
- PMCID: PMC10094455
- DOI: 10.3390/ijms24076315
Next Generation of Ovarian Cancer Detection Using Aptamers
Abstract
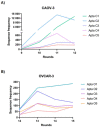
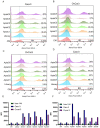
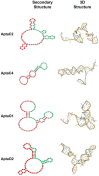
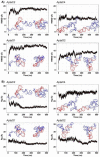
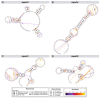
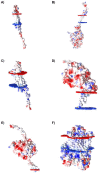
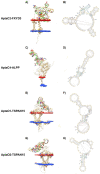
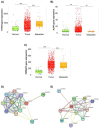
Ovarian cancer is among the seven most common types of cancer in women, being the most fatal gynecological tumor, due to the difficulty of detection in early stages. Aptamers are important tools to improve tumor diagnosis through the recognition of specific molecules produced by tumors. Here, aptamers and their potential targets in ovarian cancer cells were analyzed by in silico approaches. Specific aptamers were selected by the Cell-SELEX method using Caov-3 and OvCar-3 cells. The five most frequent aptamers obtained from the last round of selection were computationally modeled. The potential targets for those aptamers in cells were proposed by analyzing proteomic data available for the Caov-3 and OvCar-3 cell lines. Overexpressed proteins for each cell were characterized as to their three-dimensional model, cell location, and electrostatic potential. As a result, four specific aptamers for ovarian tumors were selected: AptaC2, AptaC4, AptaO1, and AptaO2. Potential targets were identified for each aptamer through Molecular Docking, and the best complexes were AptaC2-FXYD3, AptaC4-ALPP, AptaO1-TSPAN15, and AptaO2-TSPAN15. In addition, AptaC2 and AptaO1 could detect different stages and subtypes of ovarian cancer tissue samples. The application of this technology makes it possible to propose new molecular biomarkers for the differential diagnosis of epithelial ovarian cancer.
Keywords: aptamers; computational modeling; ovarian cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Mrochem J., Sodowski K., Deja R., Walaszek-Gruszka A., Wojcieszek A., Kołosza Z., Chmura A., Czernik E., Masłyk B., Bartnik W., et al. Evaluation of selected serum protein markers as early detectors of ovarian cancer. Ginekol. Pol. 2008;79:271–275. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

