Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders
- PMID: 37047372
- PMCID: PMC10094141
- DOI: 10.3390/ijms24076398
Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders
Abstract
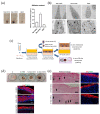
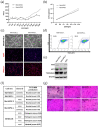
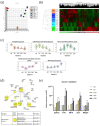
Melanocytes are essential for skin homeostasis and protection, and their loss or misfunction leads to a wide spectrum of diseases. Cell therapy utilizing autologous melanocytes has been used for years as an adjunct treatment for hypopigmentary disorders such as vitiligo. However, these approaches are hindered by the poor proliferative capacity of melanocytes obtained from skin biopsies. Recent advances in the field of human pluripotent stem cells have fueled the prospect of generating melanocytes. Here, we have developed a well-characterized method to produce a pure and homogenous population of functional and proliferative melanocytes. The genetic stability and potential transformation of melanocytes from pluripotent stem cells have been evaluated over time during the in vitro culture process. Thanks to transcriptomic analysis, the molecular signatures all along the differentiation protocol have been characterized, providing a solid basis for standardizing the protocol. Altogether, our results promise meaningful, broadly applicable, and longer-lasting advances for pigmentation disorders and open perspectives for innovative biotherapies for pigment disorders.
Keywords: human pluripotent stem cell; pigmentation-related skin disorders; skin regeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
[Melanocytes derived from pluripotent stem cells: a new biological resource for pigmentary disorders].Med Sci (Paris). 2011 Nov;27(11):935-8. doi: 10.1051/medsci/20112711007. Epub 2011 Nov 30. Med Sci (Paris). 2011. PMID: 22130018 French. No abstract available.
-
Glabrous lesional stem cells differentiated into functional melanocytes: new hope for repigmentation.J Eur Acad Dermatol Venereol. 2016 Sep;30(9):1555-60. doi: 10.1111/jdv.13686. Epub 2016 May 11. J Eur Acad Dermatol Venereol. 2016. PMID: 27538731
-
Coloring skin with pluripotent stem cells.Cell Cycle. 2011 Dec 1;10(23):3985-6. doi: 10.4161/cc.10.23.18249. Epub 2011 Dec 1. Cell Cycle. 2011. PMID: 22101274 Review. No abstract available.
-
Human-Induced Pluripotent Stem Cell‒Derived Keratinocytes, as Therapeutic Option in Vitiligo.Methods Mol Biol. 2024;2849:185-202. doi: 10.1007/7651_2023_510. Methods Mol Biol. 2024. PMID: 38189899
-
Skin pigmentation and its control: From ultraviolet radiation to stem cells.Exp Dermatol. 2021 Apr;30(4):560-571. doi: 10.1111/exd.14260. Epub 2020 Dec 24. Exp Dermatol. 2021. PMID: 33320376 Free PMC article. Review.
Cited by
-
Differentiation of cultured hair follicle neural crest stem cells into functional melanocytes.Heliyon. 2024 Aug 7;10(15):e35295. doi: 10.1016/j.heliyon.2024.e35295. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170163 Free PMC article.
References
-
- Sułkowski M., Kot M., Badyra B., Paluszkiewicz A., Płonka P.M., Sarna M., Michalczyk-Wetula D., Zucca F.A., Zecca L., Majka M. Highly Effective Protocol for Differentiation of Induced Pluripotent Stem Cells (iPS) into Melanin-Producing Cells. Int. J. Mol. Sci. 2021;22:12787. doi: 10.3390/ijms222312787. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

