Comparison between Three Different Techniques for the Detection of EGFR Mutations in Liquid Biopsies of Patients with Advanced Stage Lung Adenocarcinoma
- PMID: 37047382
- PMCID: PMC10094170
- DOI: 10.3390/ijms24076410
Comparison between Three Different Techniques for the Detection of EGFR Mutations in Liquid Biopsies of Patients with Advanced Stage Lung Adenocarcinoma
Abstract
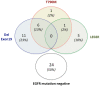
Oncogenic mutations in the EGFR gene are targets of tyrosine kinase inhibitors (TKIs) in lung adenocarcinoma (LC) patients, and their search is mandatory to make decisions on treatment strategies. Liquid biopsy of circulating tumour DNA (ctDNA) is increasingly used to detect EGFR mutations, including main activating alterations (exon 19 deletions and exon 21 L858R mutation) and T790M mutation, which is the most common mechanism of acquired resistance to first- and second-generation TKIs. In this study, we prospectively compared three different techniques for EGFR mutation detection in liquid biopsies of such patients. Fifty-four ctDNA samples from 48 consecutive advanced LC patients treated with TKIs were tested for relevant EGFR mutations with Therascreen® EGFR Plasma RGQ-PCR Kit (Qiagen). Samples were subsequently tested with two different technologies, with the aim to compare the EGFR detection rates: real-time PCR based Idylla™ ctEGFR mutation assay (Biocartis) and next-generation sequencing (NGS) system with Ion AmpliSeq Cancer Hotspot panel (ThermoFisher). A high concordance rate for main druggable EGFR alterations was observed with the two real-time PCR-based assays, ranging from 100% for T790M mutation to 94% for L858R variant and 85% for exon 19 deletions. Conversely, lower concordance rates were found between real-time PCR approaches and the NGS method (L858R: 88%; exon19-dels: 74%; T790M: 37.5%). Our results evidenced an equivalent detection ability between PCR-based techniques for circulating EGFR mutations. The NGS assay allowed detection of a wider range of EGFR mutations but showed a poor ability to detect T790M.
Keywords: EGFR; lung adenocarcinoma; mutation analysis; tyrosine kinase inhibitors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- IARC. [(accessed on 15 December 2022)]. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/15-Lung-fact-sheet.pdf.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous