Potential Use of a Combined Bacteriophage-Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study
- PMID: 37047510
- PMCID: PMC10095405
- DOI: 10.3390/ijms24076535
Potential Use of a Combined Bacteriophage-Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study
Abstract
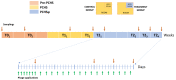
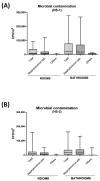
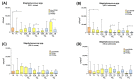
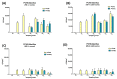
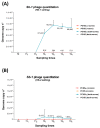
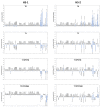
Microbial contamination in the hospital environment is a major concern for public health, since it significantly contributes to the onset of healthcare-associated infections (HAIs), which are further complicated by the alarming level of antimicrobial resistance (AMR) of HAI-associated pathogens. Chemical disinfection to control bioburden has a temporary effect and can favor the selection of resistant pathogens, as observed during the COVID-19 pandemic. Instead, probiotic-based sanitation (probiotic cleaning hygiene system, PCHS) was reported to stably abate pathogens, AMR, and HAIs. PCHS action is not rapid nor specific, being based on competitive exclusion, but the addition of lytic bacteriophages that quickly and specifically kill selected bacteria was shown to improve PCHS effectiveness. This study aimed to investigate the effect of such combined probiotic-phage sanitation (PCHSφ) in two Italian hospitals, targeting staphylococcal contamination. The results showed that PCHSφ could provide a significantly higher removal of staphylococci, including resistant strains, compared with disinfectants (-76%, p < 0.05) and PCHS alone (-50%, p < 0.05). Extraordinary sporadic chlorine disinfection appeared compatible with PCHSφ, while frequent routine chlorine usage inactivated the probiotic/phage components, preventing PCHSφ action. The collected data highlight the potential of a biological sanitation for better control of the infectious risk in healthcare facilities, without worsening pollution and AMR concerns.
Keywords: AMR; HAI; bacteriophages; bioburden; probiotics; sanitation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Effective elimination of Staphylococcal contamination from hospital surfaces by a bacteriophage-probiotic sanitation strategy: a monocentric study.Microb Biotechnol. 2019 Jul;12(4):742-751. doi: 10.1111/1751-7915.13415. Epub 2019 Apr 25. Microb Biotechnol. 2019. PMID: 31025530 Free PMC article.
-
Introduction of Probiotic-Based Sanitation in the Emergency Ward of a Children's Hospital During the COVID-19 Pandemic.Infect Drug Resist. 2022 Mar 30;15:1399-1410. doi: 10.2147/IDR.S356740. eCollection 2022. Infect Drug Resist. 2022. PMID: 35386291 Free PMC article.
-
Reducing healthcare-associated infections incidence by a probiotic-based sanitation system: A multicentre, prospective, intervention study.PLoS One. 2018 Jul 12;13(7):e0199616. doi: 10.1371/journal.pone.0199616. eCollection 2018. PLoS One. 2018. PMID: 30001345 Free PMC article.
-
Fighting AMR in the Healthcare Environment: Microbiome-Based Sanitation Approaches and Monitoring Tools.Int J Mol Sci. 2019 Mar 27;20(7):1535. doi: 10.3390/ijms20071535. Int J Mol Sci. 2019. PMID: 30934725 Free PMC article. Review.
-
Tackling transmission of infectious diseases: A probiotic-based system as a remedy for the spread of pathogenic and resistant microbes.Microb Biotechnol. 2024 Jul;17(7):e14529. doi: 10.1111/1751-7915.14529. Microb Biotechnol. 2024. PMID: 39045894 Free PMC article. Review.
Cited by
-
A New Insight into Phage Combination Therapeutic Approaches Against Drug-Resistant Mixed Bacterial Infections.Phage (New Rochelle). 2024 Dec 18;5(4):203-222. doi: 10.1089/phage.2024.0011. eCollection 2024 Dec. Phage (New Rochelle). 2024. PMID: 40045937 Review.
-
Can probiotics trigger a paradigm shift for cleaning healthcare environments? A narrative review.Antimicrob Resist Infect Control. 2024 Oct 8;13(1):119. doi: 10.1186/s13756-024-01474-6. Antimicrob Resist Infect Control. 2024. PMID: 39380032 Free PMC article. Review.
-
Profiling the resistome and virulome of Bacillus strains used for probiotic-based sanitation: a multicenter WGS analysis.BMC Genomics. 2025 Apr 18;26(1):382. doi: 10.1186/s12864-025-11582-1. BMC Genomics. 2025. PMID: 40251489 Free PMC article.
-
Biocontrol in built environments to reduce pathogen exposure and infection risk.ISME J. 2024 Jan 8;18(1):wrad024. doi: 10.1093/ismejo/wrad024. ISME J. 2024. PMID: 38365248 Free PMC article. Review.
-
Microbes Saving Lives and Reducing Suffering.Microb Biotechnol. 2025 Jan;18(1):e70068. doi: 10.1111/1751-7915.70068. Microb Biotechnol. 2025. PMID: 39844583 Free PMC article. No abstract available.
References
-
- National Academies of Sciences, Engineering and Medicine. National Academy of Engineering . Microbiomes of the Built Environment: A research Agenda for Indoor Microbiology, Human Health, and Buildings. The National Academies Press; Washington, DC, USA: 2017. p. 317. - PubMed
-
- ECDC Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals. [(accessed on 15 December 2022)];2013 Available online: https://www.ecdc.europa.eu/en/publications-data/point-prevalence-survey-....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

