Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus
- PMID: 37047525
- PMCID: PMC10095084
- DOI: 10.3390/ijms24076550
Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus
Abstract
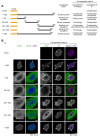
Many mononegaviruses form inclusion bodies (IBs) in infected cells. However, little is known about nuclear IBs formed by mononegaviruses, since only a few lineages of animal-derived mononegaviruses replicate in the nucleus. In this study, we characterized the IBs formed by Nyamanini virus (NYMV), a unique tick-borne mononegavirus undergoing replication in the nucleus. We discovered that NYMV forms IBs, consisting of condensates and puncta of various sizes and morphologies, in the host nucleus. Likewise, we found that the expressions of NYMV nucleoprotein (N) and phosphoprotein (P) alone induce the formation of condensates and puncta in the nucleus, respectively, even though their morphologies are somewhat different from the IBs observed in the actual NYMV-infected cells. In addition, IB-like structures can be reconstructed by co-expressions of NYMV N and P, and localization analyses using a series of truncated mutants of P revealed that the C-terminal 27 amino acid residues of P are important for recruiting P to the condensates formed by N. Furthermore, we found that nuclear speckles, cellular biomolecular condensates, are reorganized and recruited to the IB-like structures formed by the co-expressions of N and P, as well as IBs formed in NYMV-infected cells. These features are unique among mononegaviruses, and our study has contributed to elucidating the replication mechanisms of nuclear-replicating mononegaviruses and the virus-host interactions.
Keywords: Nyamanini virus; biomolecular condensates; liquid–liquid phase separation; membraneless organelles; mononegavirus; non-segmented negative-strand RNA viruses; nuclear speckles; nucleoprotein; nucleus; phosphoprotein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lahaye X., Vidy A., Pomier C., Obiang L., Harper F., Gaudin Y., Blondel D. Functional Characterization of Negri Bodies (NBs) in Rabies Virus-Infected Cells: Evidence That NBs Are Sites of Viral Transcription and Replication. J. Virol. 2009;83:7948–7958. doi: 10.1128/JVI.00554-09. - DOI - PMC - PubMed
-
- Ringel M., Heiner A., Behner L., Halwe S., Sauerhering L., Becker N., Dietzel E., Sawatsky B., Kolesnikova L., Maisner A. Nipah Virus Induces Two Inclusion Body Populations: Identification of Novel Inclusions at the Plasma Membrane. PLoS Pathog. 2019;15:e1007733. doi: 10.1371/journal.ppat.1007733. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

