Enriched Graphene Oxide-Polypropylene Suture Threads Buttons Modulate the Inflammatory Pathway Induced by Escherichia coli Lipopolysaccharide
- PMID: 37047593
- PMCID: PMC10095426
- DOI: 10.3390/ijms24076622
Enriched Graphene Oxide-Polypropylene Suture Threads Buttons Modulate the Inflammatory Pathway Induced by Escherichia coli Lipopolysaccharide
Abstract
Graphene oxide (GO), derived from graphene, has remarkable chemical-physical properties such as stability, strength, and thermal or electric conductivity and additionally shows antibacterial and anti-inflammatory properties. The present study aimed to evaluate the anti-inflammatory effects of polypropylene suture threads buttons (PPSTBs), enriched with two different concentrations of GO, in the modulation of the inflammatory pathway TLR4/MyD 88/NFκB p65/NLRP3 induced by the Escherichia coli (E. coli) lipopolysaccharide (LPS-E). The gene and the protein expression of inflammatory markers were evaluated in an in vitro model of primary human gingival fibroblasts (hGFs) by real-time PCR, western blotting, and immunofluorescence analysis. Both GO concentrations used in the polypropylene suture threads buttons-GO constructs (PPSTBs-GO) decreased the expression of inflammatory markers in hGFs treated with LPS-E. The hGFs morphology and adhesion on the PPSTBs-GO constructs were also visualized by inverted light microscopy, scanning electron microscopy (SEM), and real-time PCR. Together, these results suggest that enriched PPSTBs-GO modulates the inflammatory process through TLR4/MyD 88/NFκB p65/NLRP3 pathway.
Keywords: graphene oxide; inflammasome; inflammation; polypropylene-GO composite; suture threads.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





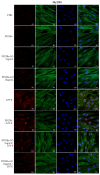




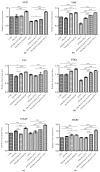

References
-
- Armano A., Agnello S. Two-Dimensional Carbon: A Review of Synthesis Methods, and Electronic, Optical, and Vibrational Properties of Single-Layer Graphene. J. Carbon Res. 2019;5:67. doi: 10.3390/c5040067. - DOI
-
- Souza I.E.P., Cambraia L.V., Gomide V.S., Nunes E.H.M. Short review on the use of graphene as a biomaterial-prospects, and challenges in Brazil. J. Mater. Res. Technol. 2022;19:2410–2430. doi: 10.1016/j.jmrt.2022.05.170. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

