Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment
- PMID: 37047624
- PMCID: PMC10095292
- DOI: 10.3390/ijms24076651
Curcumin and Plumbagin Synergistically Target the PI3K/Akt/mTOR Pathway: A Prospective Role in Cancer Treatment
Abstract
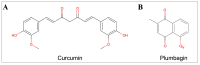
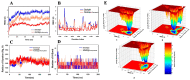
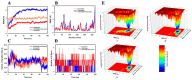
Cancer development is associated with the deregulation of various cell signaling pathways brought on by certain genetic and epigenetic alterations. Therefore, novel therapeutic strategies have been developed to target those pathways. The phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt)/mammalian target of rapamycin (mTOR) (PI3K/Akt/mTOR) pathway is one major deregulated pathway in various types of cancer. Several anticancer drug candidates are currently being investigated in preclinical and/or clinical studies to target this pathway. Natural bioactive compounds provide an excellent source for anticancer drug development. Curcumin and plumbagin are two potential anticancer compounds that have been shown to target the PI3K/Akt/mTOR pathway individually. However, their combinatorial effect on cancer cells is still unknown. This study aims to investigate the synergistic effect of these two compounds on the PI3K/Akt/mTOR pathway by employing a sequential molecular docking and molecular dynamics (MD) analysis. An increase in binding affinity and a decrease in inhibition constant have been observed when curcumin and plumbagin were subjected to sequential docking against the key proteins PI3K, Akt, and mTOR. The MD simulations and molecular mechanics combined with generalized Born surface area (MM-GBSA) analyses validated the target proteins' more stable conformation when interacting with the curcumin and plumbagin combination. This indicates the synergistic role of curcumin and plumbagin against cancer cells and the possible dose advantage when used in combination. The findings of this study pave the way for further investigation of their combinatorial effect on cancer cells in vitro and in vivo models.
Keywords: MD simulation; cancer therapy; curcumin; molecular docking; plumbagin; synergism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

