Transcriptomic Changes Predict Metabolic Alterations in LC3 Associated Phagocytosis in Aged Mice
- PMID: 37047689
- PMCID: PMC10095460
- DOI: 10.3390/ijms24076716
Transcriptomic Changes Predict Metabolic Alterations in LC3 Associated Phagocytosis in Aged Mice
Abstract
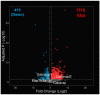
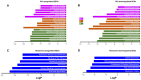
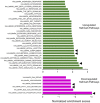
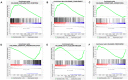
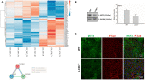
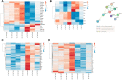
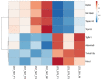
LC3b (Map1lc3b) plays an essential role in canonical autophagy and is one of several components of the autophagy machinery that mediates non-canonical autophagic functions. Phagosomes are often associated with lipidated LC3b to promote phagosome maturation in a process called LC3-associated phagocytosis (LAP). Specialized phagocytes, such as mammary epithelial cells, retinal pigment epithelial (RPE) cells, and sertoli cells, utilize LAP for optimal degradation of phagocytosed material, including debris. In the visual system, LAP is critical to maintain retinal function, lipid homeostasis, and neuroprotection. In a mouse model of retinal lipid steatosis-mice lacking LC3b (LC3b-/-), we observed increased lipid deposition, metabolic dysregulation, and enhanced inflammation. Herein, we present a non-biased approach to determine if loss of LAP mediated processes modulate the expression of various genes related to metabolic homeostasis, lipid handling, and inflammation. A comparison of the RPE transcriptome of WT and LC3b-/- mice revealed 1533 DEGs, with ~73% upregulated and 27% downregulated. Enriched gene ontology (GO) terms included inflammatory response (upregulated DEGs), fatty acid metabolism, and vascular transport (downregulated DEGs). Gene set enrichment analysis (GSEA) identified 34 pathways; 28 were upregulated (dominated by inflammation/related pathways) and 6 were downregulated (dominated by metabolic pathways). Analysis of additional gene families identified significant differences for genes in the solute carrier family, RPE signature genes, and genes with a potential role in age-related macular degeneration. These data indicate that loss of LC3b induces robust changes in the RPE transcriptome contributing to lipid dysregulation and metabolic imbalance, RPE atrophy, inflammation, and disease pathophysiology.
Keywords: LC3-associated phagocytosis (LAP); cholesterol metabolism; cholesterol trafficking; fatty acid metabolism; inflammation; monocarboxylate transporters; peroxisomes; retinal pigment epithelium (RPE); transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Update of
-
Transcriptomic changes predict metabolic alterations in LC3 associated phagocytosis in aged mice.bioRxiv [Preprint]. 2023 Mar 29:2023.03.14.532586. doi: 10.1101/2023.03.14.532586. bioRxiv. 2023. Update in: Int J Mol Sci. 2023 Apr 04;24(7):6716. doi: 10.3390/ijms24076716. PMID: 36993501 Free PMC article. Updated. Preprint.
Similar articles
-
Transcriptomic changes predict metabolic alterations in LC3 associated phagocytosis in aged mice.bioRxiv [Preprint]. 2023 Mar 29:2023.03.14.532586. doi: 10.1101/2023.03.14.532586. bioRxiv. 2023. Update in: Int J Mol Sci. 2023 Apr 04;24(7):6716. doi: 10.3390/ijms24076716. PMID: 36993501 Free PMC article. Updated. Preprint.
-
Microtubule-Associated Protein 1 Light Chain 3B, (LC3B) Is Necessary to Maintain Lipid-Mediated Homeostasis in the Retinal Pigment Epithelium.Front Cell Neurosci. 2018 Oct 8;12:351. doi: 10.3389/fncel.2018.00351. eCollection 2018. Front Cell Neurosci. 2018. PMID: 30349463 Free PMC article.
-
Peroxisome turnover and diurnal modulation of antioxidant activity in retinal pigment epithelia utilizes microtubule-associated protein 1 light chain 3B (LC3B).Am J Physiol Cell Physiol. 2019 Dec 1;317(6):C1194-C1204. doi: 10.1152/ajpcell.00185.2019. Epub 2019 Oct 2. Am J Physiol Cell Physiol. 2019. PMID: 31577510 Free PMC article.
-
Safely removing cell debris with LC3-associated phagocytosis.Biol Cell. 2017 Oct;109(10):355-363. doi: 10.1111/boc.201700028. Epub 2017 Aug 25. Biol Cell. 2017. PMID: 28755428 Review.
-
LC3-Associated Phagocytosis and Inflammation.J Mol Biol. 2017 Nov 24;429(23):3561-3576. doi: 10.1016/j.jmb.2017.08.012. Epub 2017 Aug 25. J Mol Biol. 2017. PMID: 28847720 Free PMC article. Review.
Cited by
-
How crosstalk between mitochondria, lysosomes, and other organelles can prevent or promote dry age-related macular degeneration.Exp Eye Res. 2025 Feb;251:110219. doi: 10.1016/j.exer.2024.110219. Epub 2024 Dec 22. Exp Eye Res. 2025. PMID: 39716681 Review.
References
-
- Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1) Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

