A Review of the Biosynthesis and Structural Implications of Insulin Gene Mutations Linked to Human Disease
- PMID: 37048081
- PMCID: PMC10093311
- DOI: 10.3390/cells12071008
A Review of the Biosynthesis and Structural Implications of Insulin Gene Mutations Linked to Human Disease
Abstract
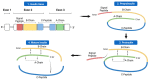
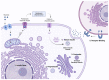
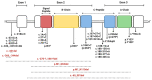
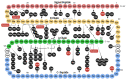
The discovery of the insulin hormone over 100 years ago, and its subsequent therapeutic application, marked a key landmark in the history of medicine and medical research. The many roles insulin plays in cell metabolism and growth have been revealed by extensive investigations into the structure and function of insulin, the insulin tyrosine kinase receptor (IR), as well as the signalling cascades, which occur upon insulin binding to the IR. In this review, the insulin gene mutations identified as causing disease and the structural implications of these mutations will be discussed. Over 100 studies were evaluated by one reviewing author, and over 70 insulin gene mutations were identified. Mutations may impair insulin gene transcription and translation, preproinsulin trafficking and proinsulin sorting, or insulin-IR interactions. A better understanding of insulin gene mutations and the resultant pathophysiology can give essential insight into the molecular mechanisms underlying impaired insulin biosynthesis and insulin-IR interaction.
Keywords: insulin biosynthesis; insulin gene mutations; mutant insulin; neonatal diabetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

