Development of Cyclic Peptides Targeting the Epidermal Growth Factor Receptor in Mesenchymal Triple-Negative Breast Cancer Subtype
- PMID: 37048151
- PMCID: PMC10093212
- DOI: 10.3390/cells12071078
Development of Cyclic Peptides Targeting the Epidermal Growth Factor Receptor in Mesenchymal Triple-Negative Breast Cancer Subtype
Abstract
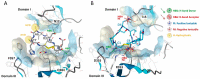
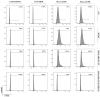
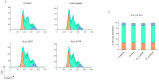
Triple-negative breast cancer (TNBC) is an aggressive malignancy characterized by the lack of expression of estrogen and progesterone receptors and amplification of human epidermal growth factor receptor 2 (HER2). Being the Epidermal Growth Factor Receptor (EGFR) highly expressed in mesenchymal TNBC and correlated with aggressive growth behavior, it represents an ideal target for anticancer drugs. Here, we have applied the phage display for selecting two highly specific peptide ligands for targeting the EGFR overexpressed in MDA-MB-231 cells, a human TNBC cell line. Molecular docking predicted the peptide-binding affinities and sites in the extracellular domain of EGFR. The binding of the FITC-conjugated peptides to human and murine TNBC cells was validated by flow cytometry. Confocal microscopy confirmed the peptide binding specificity to EGFR-positive MDA-MB-231 tumor xenograft tissues and their co-localization with the membrane EGFR. Further, the peptide stimulation did not affect the cell cycle of TNBC cells, which is of interest for their utility for tumor targeting. Our data indicate that these novel peptides are highly specific ligands for the EGFR overexpressed in TNBC cells, and thus they could be used in conjugation with nanoparticles for tumor-targeted delivery of anticancer drugs.
Keywords: EGFR; breast cancer biomarkers; peptide; phage display; triple-negative breast cancer; tumor targeting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Gupta G.K., Collier A.L., Lee D., Hoefer R.A., Zheleva V., Van Reesema L.L.S., Tang-Tan A.M., Guye M.L., Chang D.Z., Winston J.S., et al. Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers. 2020;12:2392. doi: 10.3390/cancers12092392. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

