Integrated Placental Modelling of Histology with Gene Expression to Identify Functional Impact on Fetal Growth
- PMID: 37048166
- PMCID: PMC10093760
- DOI: 10.3390/cells12071093
Integrated Placental Modelling of Histology with Gene Expression to Identify Functional Impact on Fetal Growth
Abstract
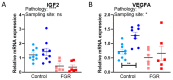
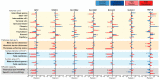
Fetal growth restriction (FGR) is a leading cause of perinatal morbidity and mortality. Altered placental formation and functional capacity are major contributors to FGR pathogenesis. Relating placental structure to function across the placenta in healthy and FGR pregnancies remains largely unexplored but could improve understanding of placental diseases. We investigated integration of these parameters spatially in the term human placenta using predictive modelling. Systematic sampling was able to overcome heterogeneity in placental morphological and molecular features. Defects in villous development, elevated fibrosis, and reduced expression of growth and functional marker genes (IGF2, VEGA, SLC38A1, and SLC2A3) were seen in age-matched term FGR versus healthy control placentas. Characteristic histopathological changes with specific accompanying molecular signatures could be integrated through computational modelling to predict if the placenta came from a healthy or FGR pregnancy. Our findings yield new insights into the spatial relationship between placental structure and function and the etiology of FGR.
Keywords: FGR; growth genes; modelling; morphology; placenta; transport.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Byrne M., Aughwane R., James J.L., Hutchinson J.C., Arthurs O.J., Sebire N.J., Ourselin S., David A.L., Melbourne A., Clark A.R. Structure-function relationships in the feto-placental circulation from in silico interpretation of micro-CT vascular structures. J. Biol. 2021;517:110630. doi: 10.1016/j.jtbi.2021.110630. - DOI - PMC - PubMed
-
- Aughwane R., Schaaf C., Hutchinson J.C., Virasami A., Zuluaga M.A., Sebire N., Arthurs O.J., Vercauteren T., Ourselin S., Melbourne A., et al. Micro-CT and histological investigation of the spatial pattern of feto-placental vascular density. Placenta. 2019;88:36–43. doi: 10.1016/j.placenta.2019.09.014. - DOI - PMC - PubMed
-
- World Health Organization . Every Newborn: An Action Plan to End Preventable Deaths. World Health Organization; Geneva, Switzerland: 2014.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

