Chinese Domestic Ducks Evolved from Mallard Duck (Anas platyrhynchos) and Spot-Billed Duck (A. zonorhyncha)
- PMID: 37048411
- PMCID: PMC10093112
- DOI: 10.3390/ani13071156
Chinese Domestic Ducks Evolved from Mallard Duck (Anas platyrhynchos) and Spot-Billed Duck (A. zonorhyncha)
Abstract
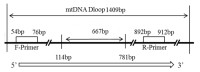
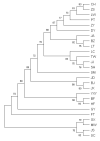
China has a rich genetic resource in its 34 domestic duck breeds. In order to detect the genetic diversity and explore the origin of these indigenous duck populations, the mitochondrial DNA (mtDNA) control region was sequenced and analyzed for 208 individual ducks, including 22 domestic breeds, wild Mallards ducks, Eastern spot-billed ducks, White Muscovy ducks, and Black Muscovy ducks. The haplotype diversity (Hd) was 0.653 and the average nucleotide diversity (Pi) was 0.005, indicating moderate genetic diversity. Sixty haplotypes were detected, and the maximum-likelihood (ML) phylogenetic tree and median-joining (MJ) network were generated from the sequence analyses. In this study, haplotypes from the Mallard duck (Anas platyrhynchos) were detected in most of the Chinese domestic duck breeds. In addition, the Eastern spot-billed duck (A. zonorhyncha) H8 haplotype was detected in two duck breeds. Only two haplotypes were found in Muscovy ducks, suggesting low genetic diversity within this population. The sequence and haplotype analyses revealed that both A. platyrhynchos and A. zonorhyncha contributed to the evolution of domestic ducks in China.
Keywords: duck; genetic diversity; mitochondrial DNA; origin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Origin and genetic diversity of Chinese domestic ducks.Mol Phylogenet Evol. 2010 Nov;57(2):634-40. doi: 10.1016/j.ympev.2010.07.011. Epub 2010 Jul 30. Mol Phylogenet Evol. 2010. PMID: 20674751
-
Genetic diversity and systematic evolution of Chinese domestic ducks along the Yangtze-Huai River.Biochem Genet. 2007 Dec;45(11-12):823-37. doi: 10.1007/s10528-007-9121-y. Epub 2007 Oct 16. Biochem Genet. 2007. PMID: 17939031
-
A homogenous nature of native Chinese duck matrilineal pool.BMC Evol Biol. 2008 Oct 29;8:298. doi: 10.1186/1471-2148-8-298. BMC Evol Biol. 2008. PMID: 18957137 Free PMC article.
-
Genetic structure of Eurasian and North American mallard ducks based on mtDNA data.Anim Genet. 2012 Jun;43(3):352-5. doi: 10.1111/j.1365-2052.2011.02248.x. Epub 2011 Sep 23. Anim Genet. 2012. PMID: 22486512
-
Vaccination of domestic ducks against H5N1 HPAI: a review.Virus Res. 2013 Dec 5;178(1):21-34. doi: 10.1016/j.virusres.2013.07.012. Epub 2013 Aug 3. Virus Res. 2013. PMID: 23916865 Review.
Cited by
-
Analysis of the Mitochondrial COI Gene and Genetic Diversity of Endangered Goose Breeds.Genes (Basel). 2024 Aug 6;15(8):1037. doi: 10.3390/genes15081037. Genes (Basel). 2024. PMID: 39202396 Free PMC article.
-
Molecular genetic characterization and meat-use functional gene identification in Jianshui yellow-brown ducks through combined resequencing and transcriptome analysis.Front Vet Sci. 2023 Dec 12;10:1269904. doi: 10.3389/fvets.2023.1269904. eCollection 2023. Front Vet Sci. 2023. PMID: 38179331 Free PMC article.
-
Optimization of pulsed electric fields-assisted thawing process conditions and its effect on the quality of Zhijiang duck meat.Food Chem X. 2024 Sep 2;24:101812. doi: 10.1016/j.fochx.2024.101812. eCollection 2024 Dec 30. Food Chem X. 2024. PMID: 39290748 Free PMC article.
-
Whole-genome selection signature differences between Chaohu and Ji'an red ducks.BMC Genomics. 2024 May 27;25(1):522. doi: 10.1186/s12864-024-10339-6. BMC Genomics. 2024. PMID: 38802792 Free PMC article.
References
-
- Feng P., Zeng T., Yang H., Chen G., Du J., Chen L., Shen J., Tao Z., Wang P., Yang L., et al. Whole-genome resequencing provides insights into the population structure and domestication signatures of ducks in eastern China. BMC Genom. 2021;22:401. doi: 10.1186/s12864-021-07710-2. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

