Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics
- PMID: 37048725
- PMCID: PMC10095019
- DOI: 10.3390/jcm12072645
Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics
Abstract
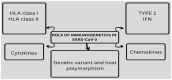
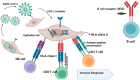
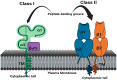
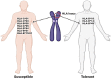
The genetic variants of HLAs (human leukocyte antigens) play a crucial role in the virus-host interaction and pathology of COVID-19. The genetic variants of HLAs not only influence T cell immune responses but also B cell immune responses by presenting a variety of peptide fragments of invading pathogens. Peptide cocktail vaccines produced by using various conserved HLA-A2 epitopes provoke substantial specific CD8+ T cell responses in experimental animals. The HLA profiles vary among individuals and trigger different T cell-mediated immune responses in COVID-19 infections. Those with HLA-C*01 and HLA-B*44 are highly susceptible to the disease. However, HLA-A*02:01, HLA-DR*03:01, and HLA-Cw*15:02 alleles show resistance to SARS infection. Understanding the genetic association of HLA with COVID-19 susceptibility and severity is important because it can help in studying the transmission of COVID-19 and its physiopathogenesis. The HLA-C*01 and B*44 allele pathways can be studied to gain insight into disease transmission and physiopathogenesis. Therefore, integrating HLA testing is suggested in the ongoing pandemic, which will help in the rapid identification of highly susceptible populations worldwide and possibly acclimate vaccine development. Therefore, understanding the correlation between HLA and SARS-CoV-2 is critical in opening new insights into COVID-19 therapeutics, based on previous studies conducted.
Keywords: ACE-2; COVID-19; HLA; antiviral immunity; disease severity; genetic susceptibility; pandemic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Does immune recognition of SARS-CoV2 epitopes vary between different ethnic groups?Virus Res. 2021 Nov;305:198579. doi: 10.1016/j.virusres.2021.198579. Epub 2021 Sep 21. Virus Res. 2021. PMID: 34560183 Free PMC article.
-
COVID-19 coronavirus vaccine T cell epitope prediction analysis based on distributions of HLA class I loci (HLA-A, -B, -C) across global populations.Hum Vaccin Immunother. 2021 Apr 3;17(4):1097-1108. doi: 10.1080/21645515.2020.1823777. Epub 2020 Nov 11. Hum Vaccin Immunother. 2021. PMID: 33175614 Free PMC article.
-
SARS-CoV-2-specific CD8+ T-cell responses and TCR signatures in the context of a prominent HLA-A*24:02 allomorph.Immunol Cell Biol. 2021 Oct;99(9):990-1000. doi: 10.1111/imcb.12482. Epub 2021 Jun 30. Immunol Cell Biol. 2021. PMID: 34086357 Free PMC article.
-
Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country.Front Immunol. 2020 Sep 3;11:2008. doi: 10.3389/fimmu.2020.02008. eCollection 2020. Front Immunol. 2020. PMID: 33013857 Free PMC article. Review.
-
Association of HLA gene polymorphism with susceptibility, severity, and mortality of COVID-19: A systematic review.HLA. 2022 Apr;99(4):281-312. doi: 10.1111/tan.14560. Epub 2022 Feb 15. HLA. 2022. PMID: 35067002
Cited by
-
The Complex Association between COPD and COVID-19.J Clin Med. 2023 May 31;12(11):3791. doi: 10.3390/jcm12113791. J Clin Med. 2023. PMID: 37297985 Free PMC article. Review.
-
The most common skin symptoms in young adults and adults related to SARS-CoV-2 virus infection.Arch Dermatol Res. 2024 May 31;316(6):292. doi: 10.1007/s00403-024-02991-5. Arch Dermatol Res. 2024. PMID: 38819524 Free PMC article. Review.
-
The role of inflammatory gene polymorphisms in severe COVID-19: a review.Virol J. 2024 Dec 20;21(1):327. doi: 10.1186/s12985-024-02597-3. Virol J. 2024. PMID: 39707400 Free PMC article. Review.
-
Revolutionizing Treatment Strategies for Autoimmune and Inflammatory Disorders: The Impact of Dipeptidyl-Peptidase 4 Inhibitors.J Inflamm Res. 2024 Mar 23;17:1897-1917. doi: 10.2147/JIR.S442106. eCollection 2024. J Inflamm Res. 2024. PMID: 38544813 Free PMC article. Review.
References
-
- Ul Haq I., Krukiewicz K., Yahya G., Haq M.U., Maryam S., Mosbah R.A., Saber S., Alrouji M. The breadth of bacteriophages contributing to the development of the phage-based vaccines for COVID-19: An ideal platform to design the multiplex vaccine. Int. J. Mol. Sci. 2023;24:1536. doi: 10.3390/ijms24021536. - DOI - PMC - PubMed
-
- Egbuna C., Chandra S., Awuchi C.G., Aklani S., Ulhaq I., Akram M., Khan J. Coronavirus Drug Discovery. Elsevier; Amsterdam, The Netherlands: 2022. Myth surrounding the FDA disapproval ofhydroxychloroquine sulfate and chloroquine phosphate as drugs for coronavirus disease 2019; pp. 153–168.
-
- Ulhaq I., Basit A., Ali I., Hussain F., Ali Z., Siddique F., Ahmed H., Aqib A.I., Rahim K. COVID-19: Epidemiology, Biochemistry, and Diagnostics. Volume 1. Bentham Science; Oak Park, IL, USA: 2021. Coronavirus Disease-2019 (COVID-19) Epidemiology; pp. 1–41. (Coronavirus Disease-19(COVID-19): A Perspective of New Scenario Series). - DOI
-
- Basit A., Ulhaq I., Hussain F., Ud-Din Z., Rahim K. COVID-19: Epidemiology, Biochemistry, and Diagnostics. Volume 1. Bentham Science; Oak Park, IL, USA: 2021. Nucleic Acid Based Detection of COVID-19; pp. 405–419. (Coronavirus Disease-19(COVID-19): A Perspective of New Scenario Series). - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

