Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach
- PMID: 37049868
- PMCID: PMC10095864
- DOI: 10.3390/molecules28073103
Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach
Abstract
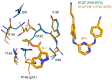
Human immunodeficiency virus type I (HIV-1) is a retrovirus that infects cells of the host's immune system leading to acquired immunodeficiency syndrome and potentially death. Although treatments are available to prevent its progression, HIV-1 remains a major burden on health resources worldwide. Continued emergence of drug-resistance mutations drives the need for novel drugs that can inhibit HIV-1 replication through new pathways. The viral protein reverse transcriptase (RT) plays a fundamental role in the HIV-1 replication cycle, and multiple approved medications target this enzyme. In this study, fragment-based drug discovery was used to optimize a previously identified hit fragment (compound B-1), which bound RT at a novel site. Three series of compounds were synthesized and evaluated for their HIV-1 RT binding and inhibition. These series were designed to investigate different vectors around the initial hit in an attempt to improve inhibitory activity against RT. Our results show that the 4-position of the core scaffold is important for binding of the fragment to RT, and a lead compound with a cyclopropyl substitution was selected and further investigated. Requirements for binding to the NNRTI-binding pocket (NNIBP) and a novel adjacent site were investigated, with lead compound 27-a minimal but efficient NNRTI-offering a starting site for the development of novel dual NNIBP-Adjacent site inhibitors.
Keywords: HIV-1; drug discovery; fragment-based drug design; non-nucleoside reverse transcriptase inhibitors (NNRTIs); reverse transcriptase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
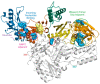
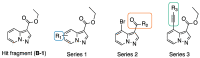
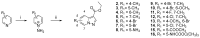
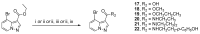
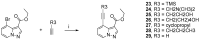


Similar articles
-
Strategies in the Design and Development of Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs).Viruses. 2023 Sep 25;15(10):1992. doi: 10.3390/v15101992. Viruses. 2023. PMID: 37896769 Free PMC article. Review.
-
Identification of "dual-site"-binding diarylpyrimidines targeting both NNIBP and the NNRTI adjacent site of the HIV-1 reverse transcriptase.Eur J Med Chem. 2023 Feb 5;247:115045. doi: 10.1016/j.ejmech.2022.115045. Epub 2022 Dec 23. Eur J Med Chem. 2023. PMID: 36577216
-
Novel broad-spectrum thiourea non-nucleoside inhibitors for the prevention of mucosal HIV transmission.Curr HIV Res. 2006 Jul;4(3):329-45. doi: 10.2174/157016206777709519. Curr HIV Res. 2006. PMID: 16842085 Review.
-
Novel tight binding PETT, HEPT and DABO-based non-nucleoside inhibitors of HIV-1 reverse transcriptase.J Enzyme Inhib Med Chem. 2006 Aug;21(4):329-50. doi: 10.1080/14756360600774413. J Enzyme Inhib Med Chem. 2006. PMID: 17059165 Review.
-
Current status of the non-nucleoside reverse transcriptase inhibitors of human immunodeficiency virus type 1.Curr Top Med Chem. 2004;4(9):921-44. doi: 10.2174/1568026043388420. Curr Top Med Chem. 2004. PMID: 15134549 Review.
Cited by
-
Antiviral activity of myricetin glycosylated compounds isolated from Marcetia taxifolia against chikungunya virus.EXCLI J. 2023 Jul 27;22:716-731. doi: 10.17179/excli2023-6242. eCollection 2023. EXCLI J. 2023. PMID: 37662709 Free PMC article.
-
The iron group transition-metal (Fe, Ru, Os) coordination of Se-doped graphitic carbon (Se@g-C3N4) nanostructures for the smart therapeutic delivery of zidovudine (ZVD) as an antiretroviral drug: a theoretical calculation perspective.RSC Adv. 2023 Nov 21;13(48):34078-34096. doi: 10.1039/d3ra06885d. eCollection 2023 Nov 16. RSC Adv. 2023. Retraction in: RSC Adv. 2025 Apr 4;15(14):10573. doi: 10.1039/d5ra90039e. PMID: 38020013 Free PMC article. Retracted.
-
Discovery of Benzisothiazolone Derivatives as Bifunctional Inhibitors of HIV-1 Reverse Transcriptase DNA Polymerase and Ribonuclease H Activities.Biomolecules. 2024 Jul 9;14(7):819. doi: 10.3390/biom14070819. Biomolecules. 2024. PMID: 39062532 Free PMC article.
References
-
- Baker L.M., Aimon A., Murray J.B., Surgenor A.E., Matassova N., Roughley S.D., Collins P.M., Krojer T., von Delft F., Hubbard R.E. Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures. Commun. Chem. 2020;3:122. doi: 10.1038/s42004-020-00367-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

