Ligand's Partition to the Lipid Bilayer Should Be Accounted for When Estimating Their Affinity to Proteins
- PMID: 37049898
- PMCID: PMC10095633
- DOI: 10.3390/molecules28073136
Ligand's Partition to the Lipid Bilayer Should Be Accounted for When Estimating Their Affinity to Proteins
Abstract
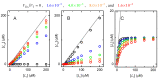
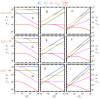
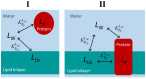
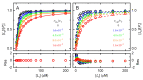
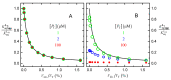
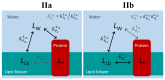
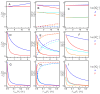
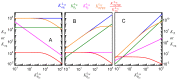
Ligand-protein interactions are usually studied in complex media that also contain lipids. This is particularly relevant for membrane proteins that are always associated with lipid bilayers, but also for water-soluble proteins studied in in vivo conditions. This work addresses the following two questions: (i) How does the neglect of the lipid bilayer influence the apparent ligand-protein affinity? (ii) How can the intrinsic ligand-protein affinity be obtained? Here we present a framework to quantitatively characterize ligand-protein interactions in complex media for proteins with a single binding site. The apparent affinity obtained when following some often-used approximations is also explored, to establish these approximations' validity limits and to allow the estimation of the true affinities from data reported in literature. It is found that an increase in the ligand lipophilicity or in the volume of the lipid bilayer always leads to a decrease in the apparent ligand-protein affinity, both for water-soluble and for membrane proteins. The only exceptions are very polar ligands (excluded from the lipid bilayer) and ligands whose binding affinity to the protein increases supralinearly with ligand lipophilicity. Finally, this work discusses which are the most relevant parameters to consider when exploring the specificity of membrane proteins.
Keywords: binding affinity; ligand exclusion; ligand sequestration; lipid-protein ratio; membrane proteins; partition coefficient; protein specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Analysis of the Equilibrium Distribution of Ligands in Heterogeneous Media-Approaches and Pitfalls.Int J Mol Sci. 2022 Aug 28;23(17):9757. doi: 10.3390/ijms23179757. Int J Mol Sci. 2022. PMID: 36077155 Free PMC article. Review.
-
Testing and characterizing enzymes and membrane-bound carrier proteins acting on amphipathic ligands in the presence of bilayer membrane material and soluble binding protein. Application to the uptake of oleate into isolated cells.Biochem J. 1992 Jun 1;284 ( Pt 2)(Pt 2):353-61. doi: 10.1042/bj2840353. Biochem J. 1992. PMID: 1599418 Free PMC article.
-
Analysis of membrane-bound acceptors. A correction function for non-specific accumulation of poorly water-soluble hydrophobic or amphipathic ligands based on the ligand partition concept.Biochem Pharmacol. 1992 Feb 18;43(4):701-4. doi: 10.1016/0006-2952(92)90233-9. Biochem Pharmacol. 1992. PMID: 1540223
-
Multivalent ligand-receptor binding on supported lipid bilayers.J Struct Biol. 2009 Oct;168(1):90-4. doi: 10.1016/j.jsb.2009.05.010. Epub 2009 Jun 7. J Struct Biol. 2009. PMID: 19508894 Free PMC article. Review.
-
Inter-Leaflet Phospholipid Exchange Impacts the Ligand Density Available for Protein Binding at Supported Lipid Bilayers.Langmuir. 2022 Jun 7;38(22):6967-6976. doi: 10.1021/acs.langmuir.2c00526. Epub 2022 May 26. Langmuir. 2022. PMID: 35617691
Cited by
-
Interaction of Hoechst 33342 with POPC Membranes at Different pH Values.Molecules. 2023 Jul 25;28(15):5640. doi: 10.3390/molecules28155640. Molecules. 2023. PMID: 37570608 Free PMC article.
-
Improving the Accuracy of Permeability Data to Gain Predictive Power: Assessing Sources of Variability in Assays Using Cell Monolayers.Membranes (Basel). 2024 Jul 14;14(7):157. doi: 10.3390/membranes14070157. Membranes (Basel). 2024. PMID: 39057665 Free PMC article. Review.
-
Permeation of a Homologous Series of NBD-Labeled Fatty Amines through Lipid Bilayers: A Molecular Dynamics Study.Membranes (Basel). 2023 May 25;13(6):551. doi: 10.3390/membranes13060551. Membranes (Basel). 2023. PMID: 37367755 Free PMC article.
References
-
- Escriba P.V., Gonzalez-Ros J.M., Goni F.M., Kinnunen P.K.J., Vigh L., Sanchez-Magraner L., Fernandez A.M., Busquets X., Horvath I., Barcelo-Coblijn G. Membranes: A meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008;12:829–875. doi: 10.1111/j.1582-4934.2008.00281.x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

