Heart failure with preserved ejection fraction phenogroup classification using machine learning
- PMID: 37051638
- PMCID: PMC10192264
- DOI: 10.1002/ehf2.14368
Heart failure with preserved ejection fraction phenogroup classification using machine learning
Abstract
Aims: Heart failure (HF) with preserved ejection fraction (HFpEF) is a complex syndrome with a poor prognosis. Phenotyping is required to identify subtype-dependent treatment strategies. Phenotypes of Japanese HFpEF patients are not fully elucidated, whose obesity is much less than Western patients. This study aimed to reveal model-based phenomapping using unsupervised machine learning (ML) for HFpEF in Japanese patients.
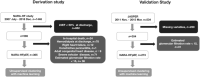
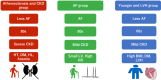
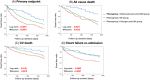
Methods and results: We studied 365 patients with HFpEF (left ventricular ejection fraction >50%) as a derivation cohort from the Nara Registry and Analyses for Heart Failure (NARA-HF), which registered patients with hospitalization by acute decompensated HF. We used unsupervised ML with a variational Bayesian-Gaussian mixture model (VBGMM) with common clinical variables. We also performed hierarchical clustering on the derivation cohort. We adopted 230 patients in the Japanese Heart Failure Syndrome with Preserved Ejection Fraction Registry as the validation cohort for VBGMM. The primary endpoint was defined as all-cause death and HF readmission within 5 years. Supervised ML was performed on the composite cohort of derivation and validation. The optimal number of clusters was three because of the probable distribution of VBGMM and the minimum Bayesian information criterion, and we stratified HFpEF into three phenogroups. Phenogroup 1 (n = 125) was older (mean age 78.9 ± 9.1 years) and predominantly male (57.6%), with the worst kidney function (mean estimated glomerular filtration rate 28.5 ± 9.7 mL/min/1.73 m2 ) and a high incidence of atherosclerotic factor. Phenogroup 2 (n = 200) had older individuals (mean age 78.8 ± 9.7 years), the lowest body mass index (BMI; 22.78 ± 3.94), and the highest incidence of women (57.5%) and atrial fibrillation (56.5%). Phenogroup 3 (n = 40) was the youngest (mean age 63.5 ± 11.2) and predominantly male (63.5 ± 11.2), with the highest BMI (27.46 ± 5.85) and a high incidence of left ventricular hypertrophy. We characterized these three phenogroups as atherosclerosis and chronic kidney disease, atrial fibrillation, and younger and left ventricular hypertrophy groups, respectively. At the primary endpoint, Phenogroup 1 demonstrated the worst prognosis (Phenogroups 1-3: 72.0% vs. 58.5% vs. 45%, P = 0.0036). We also successfully classified a derivation cohort into three similar phenogroups using VBGMM. Hierarchical and supervised clustering successfully showed the reproducibility of the three phenogroups.
Conclusions: ML could successfully stratify Japanese HFpEF patients into three phenogroups (atherosclerosis and chronic kidney disease, atrial fibrillation, and younger and left ventricular hypertrophy groups).
Keywords: Heart failure with preserved ejection fraction; Machine learning; Unsupervised clustering.
© 2023 The Authors. ESC Heart Failure published by John Wiley & Sons Ltd on behalf of European Society of Cardiology.
Conflict of interest statement
Y.S. has received research funds from Otsuka Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Bristol‐Myers Squibb Company, Actelion Pharmaceuticals Japan Ltd., Kyowa Kirin Co., Ltd., Kowa Pharmaceutical Co. Ltd., Shionogi & Co., Ltd., Dainippon Sumitomo Pharma Co., Ltd., Teijin Pharma Ltd., Chugai Pharmaceutical Co., Ltd., Eli Lilly Japan K.K., Nihon Medi‐Physics Co., Ltd., Novartis Pharma K.K., Pfizer Japan Inc., and Fuji Yakuhin Co., Ltd.; research expenses from Novartis Pharma K.K., Roche Diagnostics K.K., Amgen Inc., Bayer Yakuhin, Ltd., Astellas Pharma Inc., and Actelion Pharmaceuticals Japan Ltd.; speakers' bureau/honorarium from Alnylam Japan K.K., AstraZeneca K.K., Otsuka Pharmaceutical Co., Ltd., Kowa Pharmaceutical Co. Ltd., Daiichi Sankyo Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Tsumura & Co., Teijin Pharma Ltd., Toa Eiyo Ltd., Nippon Shinyaku Co., Ltd., Nippon Boehringer Ingelheim Co., Ltd., Novartis Pharma K.K., Bayer Yakuhin Ltd., Pfizer Japan Inc., Bristol‐Myers Squibb Company, and Mochida Pharmaceutical Co., Ltd.; and consultation fees from Ono Pharmaceutical Co., Ltd. and Novartis Pharma K.K. The remaining authors have no conflicts of interest to report.
Figures

References
-
- Owan TE, Hodge DO, Herges RM, Jacobsen SJ, Roger VL, Redfield MM. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N Engl J Med. 2006; 355: 251–259. - PubMed
-
- Anand IS, Rector TS, Cleland JG, Kuskowski M, McKelvie RS, Persson H, McMurray JJ, Zile MR, Komajda M, Massie BM, et al. Prognostic value of baseline plasma amino‐terminal pro‐brain natriuretic peptide and its interactions with irbesartan treatment effects in patients with heart failure and preserved ejection fraction findings from the I‐PRESERVE trial. Circ Heart Fail. 2011; 4: 569–577. - PubMed
-
- Anker SD, Butler J, Filippatos G, Ferreira JP, Bocchi E, Böhm M, Brunner‐La Rocca H‐P, Choi D‐J, Chopra V, Chuquiure‐Valenzuela E, et al. Empagliflozin in heart failure with a preserved ejection fraction. N Engl J Med. 2021; 385: 1451–1461. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
