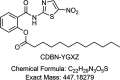
CDBN-YGXZ, a Novel Small-Molecule Drug, Shows Efficacy against Clostridioides difficile Infection and Recurrence in Mouse and Hamster Infection Models
- PMID: 37052498
- PMCID: PMC10190532
- DOI: 10.1128/aac.01704-22
CDBN-YGXZ, a Novel Small-Molecule Drug, Shows Efficacy against Clostridioides difficile Infection and Recurrence in Mouse and Hamster Infection Models
Abstract
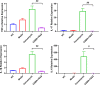
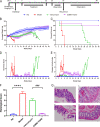
Clostridioides difficile infection (CDI) causes severe diarrhea and colitis, leading to significant morbidity, mortality, and high medical costs worldwide. Oral vancomycin, a first-line treatment for CDI, is associated with a high risk of recurrence, necessitating novel therapies for primary and recurrent CDI. A novel small-molecule compound, CDBN-YGXZ, was synthesized by modifying the benzene ring of nitazoxanide with lauric acid. The mechanism of action of CDBN-YGXZ was validated using a pyruvate:ferredoxin/flavodoxin oxidoreductase (PFOR) inhibition assay. The efficacy of CDBN-YGXZ was evaluated using the MIC test and CDI infection model in mice and hamsters. Furthermore, metagenomics was used to reveal the underlying reasons for the effective reduction or prevention of CDI after CDBN-YGXZ treatment. The inhibitory activity against PFOR induced by CDBN-YGXZ. MIC tests showed that the in vitro activity of CDBN-YGXZ against C. difficile ranging from 0.1 to 1.5 μg/mL. In the mouse and hamster CDI models, CDBN-YGXZ provided protection during both treatment and relapse, while vancomycin treatment resulted in severe relapse and significant clinical scores. Compared with global effects on the indigenous gut microbiota induced by vancomycin, CDBN-YGXZ treatment had a mild influence on gut microbes, thus resulting in the disappearance or reduction of CDI recurrence. CDBN-YGXZ displayed potent activity against C. difficile in vitro and in vivo, reducing or preventing relapse in infected animals, which could merit further development as a potential drug candidate for treating CDI.
Keywords: CDBN-YGXZ; Clostridioides difficile infection; dysbiosis; hamster; microbiome; mouse; pyruvate:ferredoxin/flavodoxin oxidoreductases; relapse/recurrence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
CDBN-YGXZ shows highly antibacterial activity against Clostridioides difficile as a potential candidate drug.Anaerobe. 2025 Jun;93:102964. doi: 10.1016/j.anaerobe.2025.102964. Epub 2025 Apr 25. Anaerobe. 2025. PMID: 40288744
-
Amixicile, a novel inhibitor of pyruvate: ferredoxin oxidoreductase, shows efficacy against Clostridium difficile in a mouse infection model.Antimicrob Agents Chemother. 2012 Aug;56(8):4103-11. doi: 10.1128/AAC.00360-12. Epub 2012 May 14. Antimicrob Agents Chemother. 2012. PMID: 22585229 Free PMC article.
-
Ridinilazole: a novel, narrow-spectrum antimicrobial agent targeting Clostridium (Clostridioides) difficile.Lett Appl Microbiol. 2022 Sep;75(3):526-536. doi: 10.1111/lam.13664. Epub 2022 Feb 11. Lett Appl Microbiol. 2022. PMID: 35119124 Free PMC article. Review.
-
A High-Carbohydrate Diet Prolongs Dysbiosis and Clostridioides difficile Carriage and Increases Delayed Mortality in a Hamster Model of Infection.Microbiol Spectr. 2022 Aug 31;10(4):e0180421. doi: 10.1128/spectrum.01804-21. Epub 2022 Jun 16. Microbiol Spectr. 2022. PMID: 35708337 Free PMC article.
-
Inhibition of spores to prevent the recurrence of Clostridioides difficile infection - A possibility or an improbability?J Microbiol Immunol Infect. 2021 Dec;54(6):1011-1017. doi: 10.1016/j.jmii.2021.06.002. Epub 2021 Jun 26. J Microbiol Immunol Infect. 2021. PMID: 34229970 Review.
Cited by
-
The state of play of rodent models for the study of Clostridioides difficile infection.J Med Microbiol. 2024 Jul;73(7):001857. doi: 10.1099/jmm.0.001857. J Med Microbiol. 2024. PMID: 39028257 Free PMC article. Review.
References
-
- Coignard B, Barbut F, Blanckaert K, Thiolet JM, Poujol I, Carbonne A, Petit JC, Desenclos JC. 2006. Emergence of Clostridium difficile toxinotype III, PCR-ribotype 027-associated disease, France, 2006. Euro Surveill 11:E060914. - PubMed
-
- Vanny PA, Wippel C, DE Andrade J, Machiavelli A, Zárate-Bladés C, Pinto A, Zamparette C, Tartari D, Sincero ARP. 2019. Therapeutic use of commensal microbes: fecal/gut microbiota transplantation. Front Drug Clin Res 5:41–110..
-
- Howerton A, Seymour CO, Murugapiran SK, Liao Z, Phan JR, Estrada A, Wagner AJ, Mefferd CC, Hedlund BP, Abel-Santos E. 2018. Effect of the synthetic bile salt analog CamSA on the hamster model of Clostridium difficile infection. Antimicrob Agents Chemother 62. doi:10.1128/AAC.02251-17. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

