Structure-aware protein self-supervised learning
- PMID: 37052532
- PMCID: PMC10139775
- DOI: 10.1093/bioinformatics/btad189
Structure-aware protein self-supervised learning
Abstract
Motivation: Protein representation learning methods have shown great potential to many downstream tasks in biological applications. A few recent studies have demonstrated that the self-supervised learning is a promising solution to addressing insufficient labels of proteins, which is a major obstacle to effective protein representation learning. However, existing protein representation learning is usually pretrained on protein sequences without considering the important protein structural information.
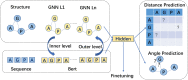
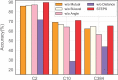
Results: In this work, we propose a novel structure-aware protein self-supervised learning method to effectively capture structural information of proteins. In particular, a graph neural network model is pretrained to preserve the protein structural information with self-supervised tasks from a pairwise residue distance perspective and a dihedral angle perspective, respectively. Furthermore, we propose to leverage the available protein language model pretrained on protein sequences to enhance the self-supervised learning. Specifically, we identify the relation between the sequential information in the protein language model and the structural information in the specially designed graph neural network model via a novel pseudo bi-level optimization scheme. We conduct experiments on three downstream tasks: the binary classification into membrane/non-membrane proteins, the location classification into 10 cellular compartments, and the enzyme-catalyzed reaction classification into 384 EC numbers, and these experiments verify the effectiveness of our proposed method.
Availability and implementation: The Alphafold2 database is available in https://alphafold.ebi.ac.uk/. The PDB files are available in https://www.rcsb.org/. The downstream tasks are available in https://github.com/phermosilla/IEConv\_proteins/tree/master/Datasets. The code of the proposed method is available in https://github.com/GGchen1997/STEPS_Bioinformatics.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Financial Support: None declared.
Figures

References
-
- Almagro Armenteros JJ, Sónderby CK, Sónderby SK. et al. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 2017;33:4049. - PubMed
-
- Anfinsen CB. Principles that govern the folding of protein chains. Science 1973;181:223–30. - PubMed
-
- Bepler T, Berger B. Learning protein sequence embeddings using information from structure. In: ICLR. 2019.
-
- Callaway E. Revolutionary cryo-EM is taking over structural biology. Nature 2020;578:201. - PubMed

