Interaction Metabolomics to Discover Synergists in Natural Product Mixtures
- PMID: 37052585
- PMCID: PMC10152448
- DOI: 10.1021/acs.jnatprod.2c00518
Interaction Metabolomics to Discover Synergists in Natural Product Mixtures
Abstract
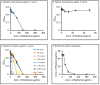
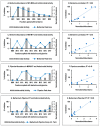
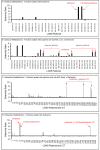
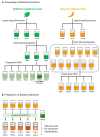
Mass spectrometry metabolomics has become increasingly popular as an integral aspect of studies to identify active compounds from natural product mixtures. Classical metabolomics data analysis approaches do not consider the possibility that interactions (such as synergy) could occur between mixture components. With this study, we developed "interaction metabolomics" to overcome this limitation. The innovation of interaction metabolomics is the inclusion of compound interaction terms (CITs), which are calculated as the product of the intensities of each pair of features (detected ions) in the data matrix. Herein, we tested the utility of interaction metabolomics by spiking known concentrations of an antimicrobial compound (berberine) and a synergist (piperine) into a set of inactive matrices. We measured the antimicrobial activity for each of the resulting mixtures against Staphylococcus aureus and analyzed the mixtures with liquid chromatography coupled to high-resolution mass spectrometry. When the data set was processed without CITs (classical metabolomics), statistical analysis yielded a pattern of false positives. However, interaction metabolomics correctly identified berberine and piperine as the compounds responsible for the synergistic activity. To further validate the interaction metabolomics approach, we prepared mixtures from extracts of goldenseal (Hydrastis canadensis) and habañero pepper (Capsicum chinense) and correctly correlated synergistic activity of these mixtures to the combined action of berberine and several capsaicinoids. Our results demonstrate the utility of a conceptually new approach for identifying synergists in mixtures that may be useful for applications in natural products research and other research areas that require comprehensive mixture analysis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Junio H. A.; Sy-Cordero A. A.; Ettefagh K. A.; Burns J. T.; Micko K. T.; Graf T. N.; Richter S. J.; Cannon R. E.; Oberlies N. H.; Cech N. B. Synergy-Directed Fractionation of Botanical Medicines: A Case Study with Goldenseal (Hydrastis Canadensis). J. Nat. Prod. 2011, 74 (7), 1621–1629. 10.1021/np200336g. - DOI - PMC - PubMed
-
- Kellogg J. J.; Todd D. A.; Egan J. M.; Raja H. A.; Oberlies N. H.; Kvalheim O. M.; Cech N. B. Biochemometrics for Natural Products Research: Comparison of Data Analysis Approaches and Application to Identification of Bioactive Compounds. J. Nat. Prod. 2016, 79 (2), 376–386. 10.1021/acs.jnatprod.5b01014. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

