Multiomics of Bohring-Opitz syndrome truncating ASXL1 mutations identify canonical and noncanonical Wnt signaling dysregulation
- PMID: 37053013
- PMCID: PMC10322691
- DOI: 10.1172/jci.insight.167744
Multiomics of Bohring-Opitz syndrome truncating ASXL1 mutations identify canonical and noncanonical Wnt signaling dysregulation
Abstract
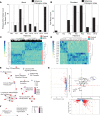
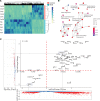
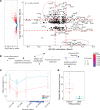
ASXL1 (additional sex combs-like 1) plays key roles in epigenetic regulation of early developmental gene expression. De novo protein-truncating mutations in ASXL1 cause Bohring-Opitz syndrome (BOS; OMIM #605039), a rare neurodevelopmental condition characterized by severe intellectual disabilities, distinctive facial features, hypertrichosis, increased risk of Wilms tumor, and variable congenital anomalies, including heart defects and severe skeletal defects giving rise to a typical BOS posture. These BOS-causing ASXL1 variants are also high-prevalence somatic driver mutations in acute myeloid leukemia. We used primary cells from individuals with BOS (n = 18) and controls (n = 49) to dissect gene regulatory changes caused by ASXL1 mutations using comprehensive multiomics assays for chromatin accessibility (ATAC-seq), DNA methylation, histone methylation binding, and transcriptome in peripheral blood and skin fibroblasts. Our data show that regardless of cell type, ASXL1 mutations drive strong cross-tissue effects that disrupt multiple layers of the epigenome. The data showed a broad activation of canonical Wnt signaling at the transcriptional and protein levels and upregulation of VANGL2, which encodes a planar cell polarity pathway protein that acts through noncanonical Wnt signaling to direct tissue patterning and cell migration. This multiomics approach identifies the core impact of ASXL1 mutations and therapeutic targets for BOS and myeloid leukemias.
Keywords: Development; Epigenetics; Genetic diseases; Genetics; Leukemias.
Conflict of interest statement
Figures


Similar articles
-
ASXL1 truncating variants in BOS and myeloid leukemia drive shared disruption of Wnt-signaling pathways but have differential isoform usage of RUNX3.BMC Med Genomics. 2024 Nov 29;17(1):282. doi: 10.1186/s12920-024-02039-7. BMC Med Genomics. 2024. PMID: 39614348 Free PMC article.
-
Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance.Am J Med Genet A. 2015 Sep;167A(9):2122-31. doi: 10.1002/ajmg.a.37131. Epub 2015 Apr 29. Am J Med Genet A. 2015. PMID: 25921057 Free PMC article.
-
Screening of CD96 and ASXL1 in 11 patients with Opitz C or Bohring-Opitz syndromes.Am J Med Genet A. 2016 Jan;170A(1):24-31. doi: 10.1002/ajmg.a.37418. Epub 2015 Oct 7. Am J Med Genet A. 2016. PMID: 26768331
-
Bohring-Opitz Syndrome.2018 Feb 15. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2018 Feb 15. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 29446906 Free Books & Documents. Review.
-
Bohring-Opitz syndrome (BOS) with a new ASXL1 pathogenic variant: Review of the most prevalent molecular and phenotypic features of the syndrome.Am J Med Genet A. 2015 Dec;167A(12):3161-6. doi: 10.1002/ajmg.a.37342. Epub 2015 Sep 14. Am J Med Genet A. 2015. PMID: 26364555 Review.
Cited by
-
Examining the neurodevelopmental and motor phenotypes of Bohring-Opitz syndrome (ASXL1) and Bainbridge-Ropers syndrome (ASXL3).Front Neurosci. 2023 Nov 6;17:1244176. doi: 10.3389/fnins.2023.1244176. eCollection 2023. Front Neurosci. 2023. PMID: 38027485 Free PMC article.
-
ASXL1 truncating variants in BOS and myeloid leukemia drive shared disruption of Wnt-signaling pathways but have differential isoform usage of RUNX3.BMC Med Genomics. 2024 Nov 29;17(1):282. doi: 10.1186/s12920-024-02039-7. BMC Med Genomics. 2024. PMID: 39614348 Free PMC article.
-
Selecting variant masks to improve power and replicability of gene-level burden tests.Res Sq [Preprint]. 2025 Apr 15:rs.3.rs-6322956. doi: 10.21203/rs.3.rs-6322956/v1. Res Sq. 2025. PMID: 40321767 Free PMC article. Preprint.
-
Asxl1 loss in mice leads to microcephaly by regulating neural stem cell survival.Anim Cells Syst (Seoul). 2025 Apr 23;29(1):241-250. doi: 10.1080/19768354.2025.2481979. eCollection 2025. Anim Cells Syst (Seoul). 2025. PMID: 40276524 Free PMC article.
-
Uncovering genomic diversity and signatures of selection in red Angus × Chinese red steppe crossbred cattle population.Sci Rep. 2025 Apr 15;15(1):12977. doi: 10.1038/s41598-025-98346-9. Sci Rep. 2025. PMID: 40234714 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

