Direct and culture-enriched 16S rRNA sequencing of cecal content of healthy horses and horses with typhlocolitis
- PMID: 37053174
- PMCID: PMC10101396
- DOI: 10.1371/journal.pone.0284193
Direct and culture-enriched 16S rRNA sequencing of cecal content of healthy horses and horses with typhlocolitis
Abstract
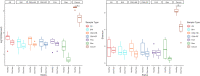
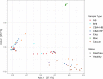
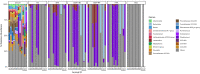
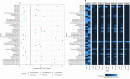
Next generation sequencing has demonstrated that alpha diversity of the fecal microbiota is significantly altered in horses with typhlocolitis. The objective of this study was to evaluate the bacterial composition of the cecum content of horses with and without typhlocolitis through direct and culture-enriched 16S gene sequencing of six healthy horses and six horses with acute typhlocolitis; a case-control study design. Cecal content was collected after euthanasia. An aliquot was used for direct 16S gene sequencing. Another was serially diluted with brain heart infusion (BHI) and plated onto five different agar media. All culture medias, except for MacConkey, were incubated anaerobically. Bacterial colonies were harvested in bulk and used for DNA extraction, 16S PCR amplification, and sequenced using the Illumina MiSeq platform. Predominant phyla in healthy and diseased horses were Firmicutes, followed by Bacteroidetes in all cultured medias, except for MacConkey agar, in which Proteobacteria was the dominant phylum. Greater bacterial richness was identified in sequenced cecal contents as compared to cultured plates (P < 0.05). Culture-enriched molecular profiling combined with 16S rRNA gene sequencing offer an alternative method for the study of the gut microbiota of horses. For direct cecum content 16S gene amplification, the alpha diversity indices were lower in diarrheic horses compared to healthy horses (P < 0.05). A higher relative abundance of Fusobacteriota was found in 2/6 samples from diarrheic horses. The role of Fusobacteriota in equine colitis deserves investigation.
Copyright: © 2023 Zakia et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Effect of hay type on cecal and fecal microbiome and fermentation parameters in horses.J Anim Sci. 2021 Jan 1;99(1):skaa407. doi: 10.1093/jas/skaa407. J Anim Sci. 2021. PMID: 33515482 Free PMC article.
-
High-throughput sequencing technology to reveal the composition and function of cecal microbiota in Dagu chicken.BMC Microbiol. 2016 Nov 4;16(1):259. doi: 10.1186/s12866-016-0877-2. BMC Microbiol. 2016. PMID: 27814685 Free PMC article.
-
Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons.FEMS Microbiol Lett. 2012 Jan;326(1):62-8. doi: 10.1111/j.1574-6968.2011.02434.x. Epub 2011 Nov 2. FEMS Microbiol Lett. 2012. PMID: 22092776
-
The Fecal Bacterial Microbiota in Horses with Equine Recurrent Uveitis.Animals (Basel). 2021 Mar 9;11(3):745. doi: 10.3390/ani11030745. Animals (Basel). 2021. PMID: 33803123 Free PMC article.
-
Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling.Genome Med. 2016 Jul 1;8(1):72. doi: 10.1186/s13073-016-0327-7. Genome Med. 2016. PMID: 27363992 Free PMC article.
Cited by
-
Vaginal and Uterine Microbiota of Healthy Maiden Mares during Estrus.Vet Sci. 2024 Jul 18;11(7):323. doi: 10.3390/vetsci11070323. Vet Sci. 2024. PMID: 39058007 Free PMC article.
-
Fecal microbiota changes associated with pathogenic and non-pathogenic diarrheas in foals.BMC Res Notes. 2025 Jan 23;18(1):34. doi: 10.1186/s13104-025-07110-9. BMC Res Notes. 2025. PMID: 39849534 Free PMC article.
-
Relationship between the components of mare breast milk and foal gut microbiome: shaping gut microbiome development after birth.Vet Q. 2024 Dec;44(1):1-9. doi: 10.1080/01652176.2024.2349948. Epub 2024 May 10. Vet Q. 2024. PMID: 38733121 Free PMC article.
-
Current Understanding of Equine Gut Dysbiosis and Microbiota Manipulation Techniques: Comparison with Current Knowledge in Other Species.Animals (Basel). 2024 Feb 28;14(5):758. doi: 10.3390/ani14050758. Animals (Basel). 2024. PMID: 38473143 Free PMC article. Review.
-
Stability of Gastric Fluid and Fecal Microbial Populations in Healthy Horses under Pasture and Stable Conditions.Animals (Basel). 2024 Oct 16;14(20):2979. doi: 10.3390/ani14202979. Animals (Basel). 2024. PMID: 39457909 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

