TFBMiner: A User-Friendly Command Line Tool for the Rapid Mining of Transcription Factor-Based Biosensors
- PMID: 37053505
- PMCID: PMC10204090
- DOI: 10.1021/acssynbio.2c00679
TFBMiner: A User-Friendly Command Line Tool for the Rapid Mining of Transcription Factor-Based Biosensors
Abstract
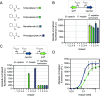
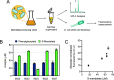
Transcription factors responsive to small molecules are essential elements in synthetic biology designs. They are often used as genetically encoded biosensors with applications ranging from the detection of environmental contaminants and biomarkers to microbial strain engineering. Despite our efforts to expand the space of compounds that can be detected using biosensors, the identification and characterization of transcription factors and their corresponding inducer molecules remain labor- and time-intensive tasks. Here, we introduce TFBMiner, a new data mining and analysis pipeline that enables the automated and rapid identification of putative metabolite-responsive transcription factor-based biosensors (TFBs). This user-friendly command line tool harnesses a heuristic rule-based model of gene organization to identify both gene clusters involved in the catabolism of user-defined molecules and their associated transcriptional regulators. Ultimately, biosensors are scored based on how well they fit the model, providing wet-lab scientists with a ranked list of candidates that can be experimentally tested. We validated the pipeline using a set of molecules for which TFBs have been reported previously, including sensors responding to sugars, amino acids, and aromatic compounds, among others. We further demonstrated the utility of TFBMiner by identifying a biosensor for S-mandelic acid, an aromatic compound for which a responsive transcription factor had not been found previously. Using a combinatorial library of mandelate-producing microbial strains, the newly identified biosensor was able to distinguish between low- and high-producing strain candidates. This work will aid in the unraveling of metabolite-responsive microbial gene regulatory networks and expand the synthetic biology toolbox to allow for the construction of more sophisticated self-regulating biosynthetic pathways.
Keywords: bioengineering; bioinformatics; biosensor; genome mining; mandelate; transcriptional regulator.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Engineering Modular Biosensors to Confer Metabolite-Responsive Regulation of Transcription.ACS Synth Biol. 2017 Feb 17;6(2):311-325. doi: 10.1021/acssynbio.6b00184. Epub 2016 Oct 31. ACS Synth Biol. 2017. PMID: 27744683
-
Genetically encoded biosensors for microbial synthetic biology: From conceptual frameworks to practical applications.Biotechnol Adv. 2023 Jan-Feb;62:108077. doi: 10.1016/j.biotechadv.2022.108077. Epub 2022 Dec 9. Biotechnol Adv. 2023. PMID: 36502964 Review.
-
Transcription-Factor-based Biosensor Engineering for Applications in Synthetic Biology.ACS Synth Biol. 2021 May 21;10(5):911-922. doi: 10.1021/acssynbio.0c00252. Epub 2021 Apr 25. ACS Synth Biol. 2021. PMID: 33899477 Review.
-
[Progress in transcription factor-based metabolite biosensors].Sheng Wu Gong Cheng Xue Bao. 2021 Mar 25;37(3):911-922. doi: 10.13345/j.cjb.200641. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 33783157 Review. Chinese.
-
Microbial Transcription Factor-Based Biosensors: Innovations from Design to Applications in Synthetic Biology.Biosensors (Basel). 2025 Mar 31;15(4):221. doi: 10.3390/bios15040221. Biosensors (Basel). 2025. PMID: 40277535 Free PMC article. Review.
Cited by
-
Ligify: Automated Genome Mining for Ligand-Inducible Transcription Factors.ACS Synth Biol. 2024 Aug 16;13(8):2577-2586. doi: 10.1021/acssynbio.4c00372. Epub 2024 Jul 19. ACS Synth Biol. 2024. PMID: 39029917 Free PMC article.
-
In vitro transcription-based biosensing of glycolate for prototyping of a complex enzyme cascade.Synth Biol (Oxf). 2024 Sep 20;9(1):ysae013. doi: 10.1093/synbio/ysae013. eCollection 2024. Synth Biol (Oxf). 2024. PMID: 39399720 Free PMC article.
-
Microbial biosensors for diagnostics, surveillance and epidemiology: Today's achievements and tomorrow's prospects.Microb Biotechnol. 2024 Nov;17(11):e70047. doi: 10.1111/1751-7915.70047. Microb Biotechnol. 2024. PMID: 39548716 Free PMC article. Review.
-
Biological Switches: Past and Future Milestones of Transcription Factor-Based Biosensors.ACS Synth Biol. 2025 Jan 17;14(1):72-86. doi: 10.1021/acssynbio.4c00689. Epub 2024 Dec 22. ACS Synth Biol. 2025. PMID: 39709556 Review.
-
Development of an Haa1-Based Biosensor for Acetic Acid Sensing in Saccharomyces cerevisiae.Methods Mol Biol. 2024;2844:221-238. doi: 10.1007/978-1-0716-4063-0_15. Methods Mol Biol. 2024. PMID: 39068343
References
-
- Zhou S.; Yuan S.-F.; Nair P. H.; Alper H. S.; Deng Y.; Zhou J. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng. 2021, 67, 41–52. 10.1016/j.ymben.2021.05.007. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

