NOTCH pathway inactivation reprograms stem-like oral cancer cells to JAK-STAT dependent state and provides the opportunity of synthetic lethality
- PMID: 37054548
- PMCID: PMC10122064
- DOI: 10.1016/j.tranon.2023.101669
NOTCH pathway inactivation reprograms stem-like oral cancer cells to JAK-STAT dependent state and provides the opportunity of synthetic lethality
Abstract
Background: We have recently provided the evidence of interconvertible cellular states, driving non-genetic heterogeneity among stem-like oral cancer cells (oral-SLCCs). Here, NOTCH pathway-activity status is explored as one of the possible mechanisms behind this stochastic plasticity.
Methods: Oral-SLCCs were enriched in 3D-spheroids. Constitutively-active and inactive status of NOTCH pathway was achieved by genetic or pharmacological approaches. RNA sequencing and real-time PCR was performed for gene expression studies. in vitro cytotoxicity assessments were performed by AlamarBlue assay and in vivo effects were studied by xenograft growth in zebrafish embryo.
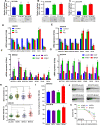
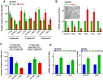
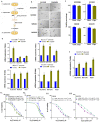
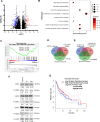
Results: We have observed stochastic plasticity in oral-SLCCs, spontaneously maintaining both NOTCH-active and inactive states. While cisplatin refraction was associated with post-treatment adaptation to the active-state of NOTCH pathway, oral-SLCCs with inactive NOTCH pathway status showed aggressive tumor growth and poor prognosis. RNAseq analysis clearly suggested the upregulation of JAK-STAT pathway in NOTCH pathway-inactive subset. The 3D-spheroids with lower NOTCH-activity status displayed significantly higher sensitivity to JAK-selective drugs, Ruxolitinib or Tofacitinib or siRNA mediated downregulation of tested partners STAT3/4. Oral-SLCCs were programmed to adapt the inactive status of NOTCH pathway by exposing to γ-secretase inhibitors, LY411575 or RO4929097, followed by targeting with JAK-inhibitors, Ruxolitinib or Tofacitinib. This approach resulted in a very significant inhibition in viability of 3D-spheroids as well as xenograft initiation in Zebrafish embryos.
Conclusion: Study revealed for the first time that NOTCH pathway-inactive state exhibit activation of JAK-STAT pathways, as synthetic lethal pair. Therefore, co-inhibition of these pathway may serve as novel therapeutic strategy against aggressive oral cancer.
Keywords: Cellular plasticity; JAK-STAT; NOTCH; Oral cancer; Stemness; Synthetic lethal pair.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors declare no potential competing interest
Figures



References
-
- Varshitha A. Prevalence of oral cancer in India. J. Pharm. Sci. Res. 2015;7(10):845–848.
-
- Mummudi N., Agarwal J.P., Chatterjee S., Mallick I., Ghosh-Laskar S. Oral cavity cancer in the indian subcontinent - challenges and opportunities. Clin. Oncol. 2019;31(8):520–528. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

