Macrophage CD5L is a target for cancer immunotherapy
- PMID: 37054630
- PMCID: PMC10139961
- DOI: 10.1016/j.ebiom.2023.104555
Macrophage CD5L is a target for cancer immunotherapy
Abstract
Background: Reprogramming of immunosuppressive tumor-associated macrophages (TAMs) presents an attractive therapeutic strategy in cancer. The aim of this study was to explore the role of macrophage CD5L protein in TAM activity and assess its potential as a therapeutic target.
Methods: Monoclonal antibodies (mAbs) against recombinant CD5L were raised by subcutaneous immunization of BALB/c mice. Peripheral blood monocytes were isolated from healthy donors and stimulated with IFN/LPS, IL4, IL10, and conditioned medium (CM) from different cancer cell lines in the presence of anti-CD5L mAb or controls. Subsequently, phenotypic markers, including CD5L, were quantified by flow cytometry, IF and RT-qPCR. Macrophage CD5L protein expression was studied in 55 human papillary lung adenocarcinoma (PAC) samples by IHC and IF. Anti-CD5L mAb and isotype control were administered intraperitoneally into a syngeneic Lewis Lung Carcinoma mouse model and tumor growth was measured. Tumor microenvironment (TME) changes were determined by flow cytometry, IHC, IF, Luminex, RNAseq and RT-qPCR.
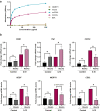
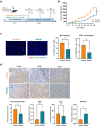
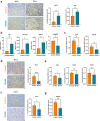
Findings: Cancer cell lines CM induced an immunosuppressive phenotype (increase in CD163, CD206, MERTK, VEGF and CD5L) in cultured macrophages. Accordingly, high TAM expression of CD5L in PAC was associated with poor patient outcome (Log-rank (Mantel-Cox) test p = 0.02). We raised a new anti-CD5L mAb that blocked the immunosuppressive phenotype of macrophages in vitro. Its administration in vivo inhibited tumor progression of lung cancer by altering the intratumoral myeloid cell population profile and CD4+ T-cell exhaustion phenotype, thereby significantly modifying the TME and increasing the inflammatory milieu.
Interpretation: CD5L protein plays a key function in modulating the activity of macrophages and their interactions within the TME, which supports its role as a therapeutic target in cancer immunotherapy.
Funding: For a full list of funding bodies, please see the Acknowledgements.
Keywords: CD5L; Immunotherapy; Lung adenocarcinoma; Macrophage; Monoclonal antibody; Scavenger receptor cysteine rich.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests A patent protecting a method for the detection of CD5L has been submitted to the European Patent Office (EP3653646A1). Likewise, the RImAb antibody is the object of an EP3476863A1 patent. LK is part of an institutional licensing agreement with SunRock Biopharma, and co-inventor of two patents (EP22382093.7 and 62828195). JB received support from MSD, Grífols and Hipra through institutional grants, and by AlbaJuna Therapeutics S.L. through an Institutional License. He is Founder and CEO of AlbaJuna Therapeutics S.L. from which he owns stock options. He is also consultant for MSD and Nesapor S.L, and received support from Gilead for attending meetings. GT has received honoraria from Takeda for lectures.
Figures





References
-
- Bejarano L., Jordāo M.J., Joyce J.A., Bejarano L., Jordāo M., Author C. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11(4):933–959. - PubMed
-
- Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17(12):887–904. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

