Parameter inference for enzyme and temperature constrained genome-scale models
- PMID: 37055413
- PMCID: PMC10102030
- DOI: 10.1038/s41598-023-32982-x
Parameter inference for enzyme and temperature constrained genome-scale models
Abstract
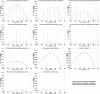
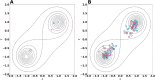
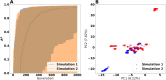
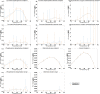
The metabolism of all living organisms is dependent on temperature, and therefore, having a good method to predict temperature effects at a system level is of importance. A recently developed Bayesian computational framework for enzyme and temperature constrained genome-scale models (etcGEM) predicts the temperature dependence of an organism's metabolic network from thermodynamic properties of the metabolic enzymes, markedly expanding the scope and applicability of constraint-based metabolic modelling. Here, we show that the Bayesian calculation method for inferring parameters for an etcGEM is unstable and unable to estimate the posterior distribution. The Bayesian calculation method assumes that the posterior distribution is unimodal, and thus fails due to the multimodality of the problem. To remedy this problem, we developed an evolutionary algorithm which is able to obtain a diversity of solutions in this multimodal parameter space. We quantified the phenotypic consequences on six metabolic network signature reactions of the different parameter solutions resulting from use of the evolutionary algorithm. While two of these reactions showed little phenotypic variation between the solutions, the remainder displayed huge variation in flux-carrying capacity. This result indicates that the model is under-determined given current experimental data and that more data is required to narrow down the model predictions. Finally, we made improvements to the software to reduce the running time of the parameter set evaluations by a factor of 8.5, allowing for obtaining results faster and with less computational resources.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Birch A N, Petersen M A, Hansen Åse S. The aroma profile of wheat bread crumb influenced by yeast concentration and fermentation temperature. LWT Food Sci. Technol. 2013;50:480–488. doi: 10.1016/j.lwt.2012.08.019. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

