Screening of Drosophila microRNA-degradation sequences reveals Argonaute1 mRNA's role in regulating miR-999
- PMID: 37055443
- PMCID: PMC10102002
- DOI: 10.1038/s41467-023-37819-9
Screening of Drosophila microRNA-degradation sequences reveals Argonaute1 mRNA's role in regulating miR-999
Abstract
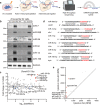
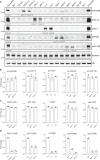
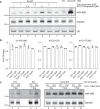
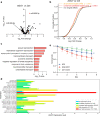
MicroRNAs (miRNA) load onto AGO proteins to target mRNAs for translational repression or degradation. However, miRNA degradation can be triggered when extensively base-paired with target RNAs, which induces confirmational change of AGO and recruitment of ZSWIM8 ubiquitin ligase to mark AGO for proteasomal degradation. This target RNA-directed miRNA degradation (TDMD) mechanism appears to be evolutionarily conserved, but recent studies have focused on mammalian systems. Here, we performed AGO1-CLASH in Drosophila S2 cells, with Dora (ortholog of vertebrate ZSWIM8) knockout mediated by CRISPR-Cas9 to identify five TDMD triggers (sequences that can induce miRNA degradation). Interestingly, one trigger in the 3' UTR of AGO1 mRNA induces miR-999 degradation. CRISPR-Cas9 knockout of the AGO1 trigger in S2 cells and in Drosophila specifically elevates miR-999, with concurrent repression of the miR-999 targets. AGO1 trigger knockout flies respond poorly to hydrogen peroxide-induced stress, demonstrating the physiological importance of this TDMD event.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
-
Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins.Genes Dev. 2021 Dec 1;35(23-24):1595-1609. doi: 10.1101/gad.348874.121. Epub 2021 Nov 24. Genes Dev. 2021. PMID: 34819352 Free PMC article.
-
The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation.Science. 2020 Dec 18;370(6523):eabc9359. doi: 10.1126/science.abc9359. Epub 2020 Nov 12. Science. 2020. PMID: 33184237 Free PMC article.
-
Insights into the target-directed miRNA degradation mechanism in Drosophila ovarian cell culture.Biochim Biophys Acta Gene Regul Mech. 2025 Jun;1868(2):195092. doi: 10.1016/j.bbagrm.2025.195092. Epub 2025 May 4. Biochim Biophys Acta Gene Regul Mech. 2025. PMID: 40328417
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
Cited by
-
Translation suppresses exogenous target RNA-mediated microRNA decay.Nat Commun. 2025 Jun 6;16(1):5257. doi: 10.1038/s41467-025-60374-4. Nat Commun. 2025. PMID: 40480978 Free PMC article.
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
-
Hybkit: a Python API and command-line toolkit for hybrid sequence data from chimeric RNA methods.Bioinformatics. 2023 Dec 1;39(12):btad721. doi: 10.1093/bioinformatics/btad721. Bioinformatics. 2023. PMID: 38006335 Free PMC article.
-
Target-directed microRNA degradation regulates developmental microRNA expression and embryonic growth in mammals.Genes Dev. 2023 Jul 1;37(13-14):661-674. doi: 10.1101/gad.350906.123. Epub 2023 Aug 8. Genes Dev. 2023. PMID: 37553261 Free PMC article.
-
Exploring new avenues of health protection: plant-derived nanovesicles reshape microbial communities.J Nanobiotechnology. 2024 May 19;22(1):269. doi: 10.1186/s12951-024-02500-w. J Nanobiotechnology. 2024. PMID: 38764018 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

