A tumor microenvironment-based prognostic index for osteosarcoma
- PMID: 37055822
- PMCID: PMC10099847
- DOI: 10.1186/s12929-023-00917-3
A tumor microenvironment-based prognostic index for osteosarcoma
Abstract
Background: The tumor microenvironment (TME) has a central role in the oncogenesis of osteosarcomas. The composition of the TME is essential for the interaction between tumor and immune cells. The aim of this study was to establish a prognostic index (TMEindex) for osteosarcoma based on the TME, from which estimates about patient survival and individual response to immune checkpoint inhibitor (ICI) therapy can be deduced.
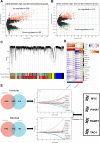
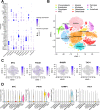
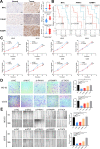
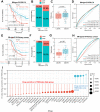
Methods: Based on osteosarcoma samples from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) database, the ESTIMATE algorithm was used to estimate ImmuneScore and StromalScore. Combined differentially expressed gene analysis, weighted gene co-expression network analyses, the Least Absolute Shrinkage and Selection Operator regression and stepwise regression to construct the TMEindex. The prognostic role of TMEindex was validated in three independent datasets. The molecular and immune characteristics of TMEindex and the impact on immunotherapy were then comprehensively investigated. The expression of TMEindex genes in different cell types and its effects on osteosarcoma cells were explored by scRNA-Seq analysis and molecular biology experiments.
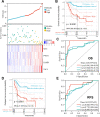
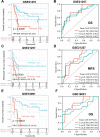
Results: Fundamental is the expression of MYC, P4HA1, RAMP1 and TAC4. Patients with high TMEindex had worse overall survival, recurrence-free survival, and metastasis-free survival. TMEindex is an independent prognostic factor in osteosarcoma. TMEindex genes were mainly expressed in malignant cells. The knockdown of MYC and P4HA1 significantly inhibited the proliferation, invasion and migration of osteosarcoma cells. A high TME index is related to the MYC, mTOR, and DNA replication-related pathways. In contrast, a low TME index is related to immune-related signaling pathways such as the inflammatory response. The TMEindex was negatively correlated with ImmuneScore, StromalScore, immune cell infiltration, and various immune-related signature scores. Patients with a higher TMEindex had an immune-cold TME and higher invasiveness. Patients with a low TME index were more likely to respond to ICI therapy and achieve clinical benefit. In addition, the TME index correlated with response to 29 oncologic drugs.
Conclusions: The TMEindex is a promising biomarker to predict the prognosis of patients with osteosarcoma and their response to ICI therapy, and to distinguish the molecular and immune characteristics.
Keywords: Big data; Checkpoint inhibitor therapy; Immune cell infiltration; Osteosarcoma; Prognostic index; Tumor microenvironment.
© 2023. The Author(s).
Conflict of interest statement
The authors declared no potential competing interests.
Figures


References
-
- Smeland S, Bielack SS, Whelan J, Bernstein M, Hogendoorn P, Krailo MD, et al. Survival and prognosis with osteosarcoma: outcomes in more than 2000 patients in the EURAMOS-1 (European and American Osteosarcoma Study) cohort. Eur J Cancer. 2019;109:36–50. doi: 10.1016/j.ejca.2018.11.027. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

