Diagnostic TR-FRET assays for detection of antibodies in patient samples
- PMID: 37056371
- PMCID: PMC10088089
- DOI: 10.1016/j.crmeth.2023.100421
Diagnostic TR-FRET assays for detection of antibodies in patient samples
Abstract
Serological assays are important diagnostic tools for surveying exposure to the pathogen, monitoring immune response post vaccination, and managing spread of the infectious agent among the population. Current serological laboratory assays are often limited because they require the use of specialized laboratory technology and/or work with a limited number of sample types. Here, we evaluate an alternative by developing time-resolved Förster resonance energy transfer (TR-FRET) homogeneous assays that exhibited exceptional versatility, scalability, and sensitivity and outperformed or matched currently used strategies in terms of sensitivity, specificity, and precision. We validated the performance of the assays measuring total immunoglobulin G (IgG) levels; antibodies against severe acute respiratory syndrome coronavirus (SARS-CoV) or Middle Eastern respiratory syndrome (MERS)-CoV spike (S) protein; and SARS-CoV-2 S and nucleocapsid (N) proteins and applied it to several large sample sets and real-world applications. We further established a TR-FRET-based ACE2-S competition assay to assess the neutralization propensity of the antibodies. Overall, these TR-FRET-based serological assays can be rapidly extended to other antigens and are compatible with commonly used plate readers.
Keywords: CoraFluor; TR-FRET assays; neutralization assay; serological testing; vaccine efficacy studies.
© 2023 The Author(s).
Conflict of interest statement
E.S.F. is an equity holder and scientific advisor for Neomorph, Inc. (board member), Civetta Therapeutics, Proximity Therapeutics, Lighthorse Therapeutics, Avilar Therapeutics, and Photys Therapeutics and is a consultant to Novartis, Sanofi, AbbVie, Pfizer, Astellas, EcoR1 Capital, and Deerfield. The Fischer lab receives or has received research funding from Novartis, Ajax, Deerfield, and Astellas not related to this work. R.M. is a scientific advisory board (SAB) member and equity holder of Regenacy Pharmaceuticals, ERX Pharmaceuticals, and Frequency Therapeutics. H.Y., R.P.N., D.O., N.C.P., R.M., and E.S.F. are inventors on patent applications related to this work.
Figures
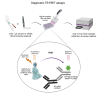


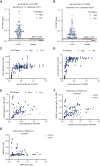
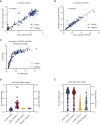


Update of
-
Rapid 'mix and read' assay for scalable detection of SARS-CoV-2 antibodies in patient plasma.medRxiv [Preprint]. 2020 Sep 3:2020.09.01.20184101. doi: 10.1101/2020.09.01.20184101. medRxiv. 2020. Update in: Cell Rep Methods. 2023 Feb 20;3(3):100421. doi: 10.1016/j.crmeth.2023.100421. PMID: 32909004 Free PMC article. Updated. Preprint.
References
-
- Corradini P., Gobbi G., de Braud F., Rosa J., Rusconi C., Apolone G., Carniti C. Rapid antibody testing for SARS-CoV-2 in asymptomatic and paucisymptomatic healthcare professionals in hematology and oncology units identifies undiagnosed infections. Hemasphere. 2020;4:e408. doi: 10.1097/HS9.0000000000000408. - DOI - PMC - PubMed
-
- Ragó Z., Szijjártó L., Duda E., Bella Z. [Opportunity of periodic monitoring of COVID-19 patients, asymptomatic virus carriers, and postinfectious individuals with IgM/IgG rapid antibody tests among healthcare workers during SARS-CoV-2 pandemic] Orv. Hetil. 2020;161:854–860. doi: 10.1556/650.2020.31862. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

