APOBEC mutational signature predicts prognosis and immunotherapy response in nonsmoking patients with lung adenocarcinoma
- PMID: 37057114
- PMCID: PMC10088004
- DOI: 10.21037/tlcr-23-150
APOBEC mutational signature predicts prognosis and immunotherapy response in nonsmoking patients with lung adenocarcinoma
Abstract
Background: Lung adenocarcinoma (LUAD) is the most common type of non-small cell lung cancer (NSCLC) with poor survival in advanced stage. Nowadays the rate of nonsmoking patients has dramatically increased and may be associated with the presence of driver mutations. Better understanding of the mutation profile data of nonsmoking LUAD patients are critical to predict survival and provide greater benefits to more patients. The apolipoprotein B mRNA editing enzyme catalytic polypeptide-like (APOBEC) has been shown to play an important role in molecular tumorigenesis of NSCLC. However, the clinical relevance of APOBEC in nonsmoking LUAD remains to be understood.
Methods: LUAD patients with somatic mutation and RNA sequencing data obtained from The Cancer Genome Atlas (TCGA) were assessed and screened in the Gene Expression Omnibus. Transcriptome data and mutational signatures were analyzed using R package. Then, we used the least absolute shrinkage and selection operator (LASSO) regression model to construct the APOBEC3 score (APOBEC3 score) model. The prognostic value was evaluated using Kaplan-Meier analysis. Finally, the functional enrichment analysis of differential expressed genes (DEGs) and the immune-related features were also estimated using R package.
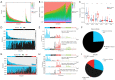
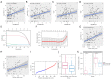
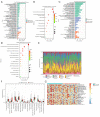
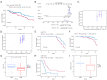
Results: By analyzing the mutational profile data of NSCLC in the TCGA database, we found that different mutation patterns existed between smoking and nonsmoking patients, and the APOBEC3 family played an important role in the mutation pattern of nonsmoking patients with LUAD. We established an APOBEC3 score and found that TCW (W = A or T) mutation counts were significantly greater in the high APOBEC3 score group than in the low APOBEC3 score group. Furthermore, there were different immune feathers and prognostic values between the high and low APOBEC3 score patients, suggesting an independent prognostic factor of APOBEC3 in nonsmoking LUAD patients.
Conclusions: We established a comprehensive view of APOBEC3 mutations in nonsmoking LUAD patients. Our review provides new insights into using the APOBEC3 mutation to predict prognosis and improve the immunotherapy response for future applications.
Keywords: Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3); immunotherapy; mutational signature; nonsmoking; prognosis.
2023 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-150/coif). The authors have no conflicts of interest to declare.
Figures

Similar articles
-
[Mutational Signatures Analysis of Micropapillary Components and Exploration of ZNF469 Gene in Early-stage Lung Adenocarcinoma with Ground-glass Opacities].Zhongguo Fei Ai Za Zhi. 2024 Jan 2;26(12):889-900. doi: 10.3779/j.issn.1009-3419.2023.106.23. Epub 2023 Dec 28. Zhongguo Fei Ai Za Zhi. 2024. PMID: 38151328 Free PMC article. Chinese.
-
Pan-cancer analysis identifies proteasome 26S subunit, ATPase (PSMC) family genes, and related signatures associated with prognosis, immune profile, and therapeutic response in lung adenocarcinoma.Front Genet. 2023 Jan 9;13:1017866. doi: 10.3389/fgene.2022.1017866. eCollection 2022. Front Genet. 2023. PMID: 36699466 Free PMC article.
-
High Expression of UBB, RAC1, and ITGB1 Predicts Worse Prognosis among Nonsmoking Patients with Lung Adenocarcinoma through Bioinformatics Analysis.Biomed Res Int. 2020 Oct 20;2020:2071593. doi: 10.1155/2020/2071593. eCollection 2020. Biomed Res Int. 2020. PMID: 33134373 Free PMC article.
-
Clinical Significance and Immunologic Landscape of a Five-IL(R)-Based Signature in Lung Adenocarcinoma.Front Immunol. 2021 Aug 23;12:693062. doi: 10.3389/fimmu.2021.693062. eCollection 2021. Front Immunol. 2021. PMID: 34497605 Free PMC article.
-
Addressing the benefits of inhibiting APOBEC3-dependent mutagenesis in cancer.Nat Genet. 2022 Nov;54(11):1599-1608. doi: 10.1038/s41588-022-01196-8. Epub 2022 Oct 24. Nat Genet. 2022. PMID: 36280735 Free PMC article. Review.
Cited by
-
FAT1 mutation-related signature predicts survival risk and tumor immunogenicity in lung adenocarcinoma.Front Genet. 2025 Jul 2;16:1466484. doi: 10.3389/fgene.2025.1466484. eCollection 2025. Front Genet. 2025. PMID: 40672387 Free PMC article.
-
[Mutational Signatures Analysis of Micropapillary Components and Exploration of ZNF469 Gene in Early-stage Lung Adenocarcinoma with Ground-glass Opacities].Zhongguo Fei Ai Za Zhi. 2024 Jan 2;26(12):889-900. doi: 10.3779/j.issn.1009-3419.2023.106.23. Epub 2023 Dec 28. Zhongguo Fei Ai Za Zhi. 2024. PMID: 38151328 Free PMC article. Chinese.
References
LinkOut - more resources
Full Text Sources
