Genotype Diversity of Enteric Viruses in Wastewater Amid the COVID-19 Pandemic
- PMID: 37058225
- PMCID: PMC10103036
- DOI: 10.1007/s12560-023-09553-4
Genotype Diversity of Enteric Viruses in Wastewater Amid the COVID-19 Pandemic
Abstract
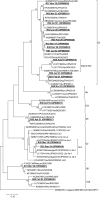
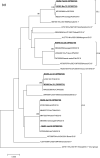
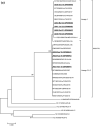
Viruses remain the leading cause of acute gastroenteritis (AGE) worldwide. Recently, we reported the abundance of AGE viruses in raw sewage water (SW) during the COVID-19 pandemic, when viral AGE patients decreased dramatically in clinics. Since clinical samples were not reflecting the actual state, it remained important to determine the circulating strains in the SW for preparedness against impending outbreaks. Raw SW was collected from a sewage treatment plant in Japan from August 2018 to March 2022, concentrated by polyethylene-glycol-precipitation method, and investigated for major gastroenteritis viruses by RT-PCR. Genotypes and evolutionary relationships were evaluated through sequence-based analyses. Major AGE viruses like rotavirus A (RVA), norovirus (NoV) GI and GII, and astrovirus (AstV) increased sharply (10-20%) in SW during the COVID-19 pandemic, though some AGE viruses like sapovirus (SV), adenovirus (AdV), and enterovirus (EV) decreased slightly (3-10%). The prevalence remained top in the winter. Importantly, several strains, including G1 and G3 of RVA, GI.1 and GII.2 of NoV, GI.1 of SV, MLB1 of AstV, and F41 of AdV, either emerged or increased amid the pandemic, suggesting that the normal phenomenon of genotype changing remained active over this time. This study crucially presents the molecular characteristics of circulating AGE viruses, explaining the importance of SW investigation during the pandemic when a clinical investigation may not produce the complete scenario.
Keywords: COVID-19 pandemic; Enteric viruses; Genotypes; Raw sewage.
© 2023. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors have no financial or proprietary interests in any material discussed in this article.
Figures




Similar articles
-
The Emergence and Widespread Circulation of Enteric Viruses Throughout the COVID-19 Pandemic: A Wastewater-Based Evidence.Food Environ Virol. 2023 Dec;15(4):342-354. doi: 10.1007/s12560-023-09566-z. Epub 2023 Oct 29. Food Environ Virol. 2023. PMID: 37898959
-
Alarming Situation of Spreading Enteric Viruses Through Sewage Water in Dhaka City: Molecular Epidemiological Evidences.Food Environ Virol. 2019 Mar;11(1):65-75. doi: 10.1007/s12560-018-09363-z. Epub 2019 Jan 3. Food Environ Virol. 2019. PMID: 30607905
-
Clinical Impact and Genetic Analysis of Enteric Viruses Associated With Acute Gastroenteritis in Greater Accra, Ghana: A Comprehensive Study of Five Viruses.J Med Virol. 2025 Feb;97(2):e70216. doi: 10.1002/jmv.70216. J Med Virol. 2025. PMID: 39935201
-
Inadequately treated wastewater as a source of human enteric viruses in the environment.Int J Environ Res Public Health. 2010 Jun;7(6):2620-37. doi: 10.3390/ijerph7062620. Epub 2010 Jun 14. Int J Environ Res Public Health. 2010. PMID: 20644692 Free PMC article. Review.
-
Extension of probability models of the risk of infections by human enteric viruses.Math Biosci Eng. 2023 Sep 13;20(9):17499-17519. doi: 10.3934/mbe.2023777. Math Biosci Eng. 2023. PMID: 37920063 Review.
Cited by
-
Assessment of Gastroenteric Viruses in Marketed Bivalve Mollusks in the Tourist Cities of Rio de Janeiro, Brazil, 2022.Viruses. 2024 Feb 20;16(3):317. doi: 10.3390/v16030317. Viruses. 2024. PMID: 38543684 Free PMC article.
-
Rotavirus in Water Environments: A Systematic Review and Meta-Analysis.Environ Health Insights. 2024 Oct 18;18:11786302241276667. doi: 10.1177/11786302241276667. eCollection 2024. Environ Health Insights. 2024. PMID: 39439598 Free PMC article. Review.
-
Metatranscriptomic identification of novel RNA viruses from raccoon dog (Nyctereutes procyonoides) feces in Japan.Sci Rep. 2025 Feb 27;15(1):7100. doi: 10.1038/s41598-025-90474-6. Sci Rep. 2025. PMID: 40016305 Free PMC article.
-
Optimal sampling frequency and site selection for wastewater and environmental surveillance of infectious pathogens: A value of information assessment.PLoS Comput Biol. 2025 Jun 25;21(6):e1013190. doi: 10.1371/journal.pcbi.1013190. eCollection 2025 Jun. PLoS Comput Biol. 2025. PMID: 40561147 Free PMC article.
-
Diverse genotypes of norovirus genogroup I and II contamination in environmental water in Thailand during the COVID-19 outbreak from 2020 to 2022.Virol Sin. 2024 Aug;39(4):556-564. doi: 10.1016/j.virs.2024.05.010. Epub 2024 May 30. Virol Sin. 2024. PMID: 38823781 Free PMC article.
References
-
- Adachi Katayama Y, Hayase S, Ando Y, Kuroita T, Okada K, Iwamoto R, et al. COPMAN: a novel high-throughput and highly sensitive method to detect viral nucleic acids including SARS-CoV-2 RNA in wastewater. Science of the Total Environment. 2023;856(Pt 1):158966. doi: 10.1016/j.scitotenv.2022.158966. - DOI - PMC - PubMed
-
- Braeckman T, Van Herck K, Meyer N, Pircon JY, Soriano-Gabarro M, Heylen E, et al. Effectiveness of rotavirus vaccination in prevention of hospital admissions for rotavirus gastroenteritis among young children in Belgium: Case-control study. BMJ. 2012;345:e4752. doi: 10.1136/bmj.e4752. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

