A Novel Patient-Personalized Nanovector Based on Homotypic Recognition and Magnetic Hyperthermia for an Efficient Treatment of Glioblastoma Multiforme
- PMID: 37058273
- PMCID: PMC11468287
- DOI: 10.1002/adhm.202203120
A Novel Patient-Personalized Nanovector Based on Homotypic Recognition and Magnetic Hyperthermia for an Efficient Treatment of Glioblastoma Multiforme
Abstract
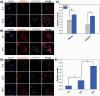
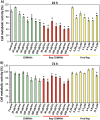
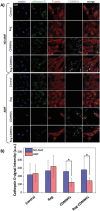
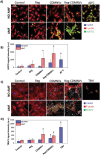
Glioblastoma multiforme (GBM) is the deadliest brain tumor, characterized by an extreme genotypic and phenotypic variability, besides a high infiltrative nature in healthy tissues. Apart from very invasive surgical procedures, to date, there are no effective treatments, and life expectancy is very limited. In this work, an innovative therapeutic approach based on lipid-based magnetic nanovectors is proposed, owning a dual therapeutic function: chemotherapy, thanks to an antineoplastic drug (regorafenib) loaded in the core, and localized magnetic hyperthermia, thanks to the presence of iron oxide nanoparticles, remotely activated by an alternating magnetic field. The drug is selected based on ad hoc patient-specific screenings; moreover, the nanovector is decorated with cell membranes derived from patients' cells, aiming at increasing homotypic and personalized targeting. It is demonstrated that this functionalization not only enhances the selectivity of the nanovectors toward patient-derived GBM cells, but also their blood-brain barrier in vitro crossing ability. The localized magnetic hyperthermia induces both thermal and oxidative intracellular stress that lead to lysosomal membrane permeabilization and to the release of proteolytic enzymes into the cytosol. Collected results show that hyperthermia and chemotherapy work in synergy to reduce GBM cell invasion properties, to induce intracellular damage and, eventually, to prompt cellular death.
Keywords: glioblastoma; homotypic targeting; magnetic hyperthermia; personalized medicine.
© 2023 The Authors. Advanced Healthcare Materials published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Stimuli-responsive lipid-based magnetic nanovectors increase apoptosis in glioblastoma cells through synergic intracellular hyperthermia and chemotherapy.Nanoscale. 2018 Dec 20;11(1):72-88. doi: 10.1039/c8nr05520c. Nanoscale. 2018. PMID: 30357214 Free PMC article.
-
Hybrid Magnetic Nanovectors Promote Selective Glioblastoma Cell Death through a Combined Effect of Lysosomal Membrane Permeabilization and Chemotherapy.ACS Appl Mater Interfaces. 2020 Jul 1;12(26):29037-29055. doi: 10.1021/acsami.0c05556. Epub 2020 Jun 8. ACS Appl Mater Interfaces. 2020. PMID: 32459082 Free PMC article.
-
Therapeutic evaluation of magnetic hyperthermia using Fe3O4-aminosilane-coated iron oxide nanoparticles in glioblastoma animal model.Einstein (Sao Paulo). 2019 Aug 1;17(4):eAO4786. doi: 10.31744/einstein_journal/2019AO4786. Einstein (Sao Paulo). 2019. PMID: 31390427 Free PMC article.
-
Evolution of Magnetic Hyperthermia for Glioblastoma Multiforme Therapy.ACS Chem Neurosci. 2019 Mar 20;10(3):1157-1172. doi: 10.1021/acschemneuro.8b00652. Epub 2019 Feb 19. ACS Chem Neurosci. 2019. PMID: 30715851 Review.
-
Nanoparticles for hyperthermic therapy: synthesis strategies and applications in glioblastoma.Int J Nanomedicine. 2014 Jun 10;9:2863-77. doi: 10.2147/IJN.S57501. eCollection 2014. Int J Nanomedicine. 2014. PMID: 24959075 Free PMC article. Review.
Cited by
-
Towards Effective Treatment of Glioblastoma: The Role of Combination Therapies and the Potential of Phytotherapy and Micotherapy.Curr Issues Mol Biol. 2024 Dec 19;46(12):14324-14350. doi: 10.3390/cimb46120859. Curr Issues Mol Biol. 2024. PMID: 39727987 Free PMC article. Review.
-
Development and characterization of lipid nanocapsules loaded with iron oxide nanoparticles for magnetic targeting to the blood-brain barrier.Drug Deliv Transl Res. 2024 Dec;14(12):3494-3511. doi: 10.1007/s13346-024-01587-w. Epub 2024 May 13. Drug Deliv Transl Res. 2024. PMID: 38739319 Free PMC article.
-
Magnetic nanomaterials for hyperthermia-based therapy and controlled drug delivery.Bioact Mater. 2025 Jul 26;53:591-629. doi: 10.1016/j.bioactmat.2025.07.033. eCollection 2025 Nov. Bioact Mater. 2025. PMID: 40761549 Free PMC article. Review.
-
Polydopamine Nanoparticle-Based Combined Chemotherapy and Photothermal Therapy for the Treatment of Liver Cancer.ACS Appl Mater Interfaces. 2024 Aug 7;16(31):40695-40713. doi: 10.1021/acsami.4c08491. Epub 2024 Jul 26. ACS Appl Mater Interfaces. 2024. PMID: 39058979 Free PMC article.
-
Cell Membrane Fragment-Wrapped Parenteral Nanoemulsions: A New Drug Delivery Tool to Target Gliomas.Cells. 2024 Apr 6;13(7):641. doi: 10.3390/cells13070641. Cells. 2024. PMID: 38607080 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

