Exercise training remodels inguinal white adipose tissue through adaptations in innervation, vascularization, and the extracellular matrix
- PMID: 37058410
- PMCID: PMC10374102
- DOI: 10.1016/j.celrep.2023.112392
Exercise training remodels inguinal white adipose tissue through adaptations in innervation, vascularization, and the extracellular matrix
Abstract
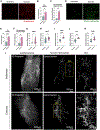
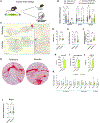
Inguinal white adipose tissue (iWAT) is essential for the beneficial effects of exercise training on metabolic health. The underlying mechanisms for these effects are not fully understood, and here, we test the hypothesis that exercise training results in a more favorable iWAT structural phenotype. Using biochemical, imaging, and multi-omics analyses, we find that 11 days of wheel running in male mice causes profound iWAT remodeling including decreased extracellular matrix (ECM) deposition and increased vascularization and innervation. We identify adipose stem cells as one of the main contributors to training-induced ECM remodeling, show that the PRDM16 transcriptional complex is necessary for iWAT remodeling and beiging, and discover neuronal growth regulator 1 (NEGR1) as a link between PRDM16 and neuritogenesis. Moreover, we find that training causes a shift from hypertrophic to insulin-sensitive adipocyte subpopulations. Exercise training leads to remarkable adaptations to iWAT structure and cell-type composition that can confer beneficial changes in tissue metabolism.
Keywords: Adipo-Clear; CP: Metabolism; CP: Molecular biology; ECM; NEGR1; PRDM16; exercise; innervation; proteomics; spatial transcriptomics; vascularization; white adipose tissue.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Nigro P, Middelbeek RJW, Alves CRR, Rovira-Llopis S, Ramachandran K, Rowland LA, Møller AB, Takahashi H, Alves-Wagner AB, Vamvini M, et al. (2021). Exercise training promotes sex-specific adaptations in mouse inguinal white adipose tissue. Diabetes 70, 1250–1264. 10.2337/db20-0790. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

