Analytical validation of a multi-cancer early detection test with cancer signal origin using a cell-free DNA-based targeted methylation assay
- PMID: 37058491
- PMCID: PMC10104288
- DOI: 10.1371/journal.pone.0283001
Analytical validation of a multi-cancer early detection test with cancer signal origin using a cell-free DNA-based targeted methylation assay
Abstract
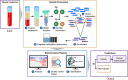
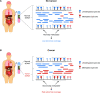
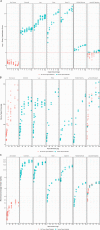
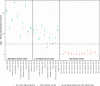
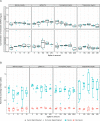
The analytical validation is reported for a targeted methylation-based cell-free DNA multi-cancer early detection test designed to detect cancer and predict the cancer signal origin (tissue of origin). A machine-learning classifier was used to analyze the methylation patterns of >105 genomic targets covering >1 million methylation sites. Analytical sensitivity (limit of detection [95% probability]) was characterized with respect to tumor content by expected variant allele frequency and was determined to be 0.07%-0.17% across five tumor cases and 0.51% for the lymphoid neoplasm case. Test specificity was 99.3% (95% confidence interval, 98.6-99.7%). In the reproducibility and repeatability study, results were consistent in 31/34 (91.2%) pairs with cancer and 17/17 (100%) pairs without cancer; between runs, results were concordant for 129/133 (97.0%) cancer and 37/37 (100%) non-cancer sample pairs. Across 3- to 100-ng input levels of cell-free DNA, cancer was detected in 157/182 (86.3%) cancer samples but not in any of the 62 non-cancer samples. In input titration tests, cancer signal origin was correctly predicted in all tumor samples detected as cancer. No cross-contamination events were observed. No potential interferent (hemoglobin, bilirubin, triglycerides, genomic DNA) affected performance. The results of this analytical validation study support continued clinical development of a targeted methylation cell-free DNA multi-cancer early detection test.
Copyright: © 2023 Alexander et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: GA, WL, FEO, MR, PS, JRB, LE, GN, AM, PWM and NH are former employees of GRAIL, LLC and may have equity in the company. MHM is a consultant to Delfi Diagnostics and to Fellow Health and a former employee of GRAIL, LLC, with equity in all three companies. AMA is an advisor to Foresite Labs, San Francisco, California and Boston, Massachusetts, and a former employee of GRAIL, LLC, and holds equity in both companies. All other authors are employees of GRAIL, LLC, with equity in the company. GRAIL, LLC is a subsidiary of Illumina, Inc. currently held separate from Illumina Inc. under the terms of the Interim Measures Order of the European Commission dated 29 October 2021.
Figures

References
-
- Siu AL U. S. Preventive Services Task Force. Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2016;164: 279–296. - PubMed
-
- Moyer VA U. S. Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160: 330–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

