Cryo-EM structure of the human Sirtuin 6-nucleosome complex
- PMID: 37058572
- PMCID: PMC10104460
- DOI: 10.1126/sciadv.adf7586
Cryo-EM structure of the human Sirtuin 6-nucleosome complex
Abstract
Sirtuin 6 (SIRT6) is a multifaceted protein deacetylase/deacylase and a major target for small-molecule modulators of longevity and cancer. In the context of chromatin, SIRT6 removes acetyl groups from histone H3 in nucleosomes, but the molecular basis for its nucleosomal substrate preference is unknown. Our cryo-electron microscopy structure of human SIRT6 in complex with the nucleosome shows that the catalytic domain of SIRT6 pries DNA from the nucleosomal entry-exit site and exposes the histone H3 N-terminal helix, while the SIRT6 zinc-binding domain binds to the histone acidic patch using an arginine anchor. In addition, SIRT6 forms an inhibitory interaction with the C-terminal tail of histone H2A. The structure provides insights into how SIRT6 can deacetylate both H3 K9 and H3 K56.
Figures







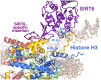
Update of
-
Cryo-EM structure of the human Sirtuin 6-nucleosome complex.bioRxiv [Preprint]. 2023 Mar 18:2023.03.17.533206. doi: 10.1101/2023.03.17.533206. bioRxiv. 2023. Update in: Sci Adv. 2023 Apr 14;9(15):eadf7586. doi: 10.1126/sciadv.adf7586. PMID: 36993468 Free PMC article. Updated. Preprint.
References
-
- R. Mostoslavsky, K. F. Chua, D. B. Lombard, W. W. Pang, M. R. Fischer, L. Gellon, P. Liu, G. Mostoslavsky, S. Franco, M. M. Murphy, K. D. Mills, P. Patel, J. T. Hsu, A. L. Hong, E. Ford, H. L. Cheng, C. Kennedy, N. Nunez, R. Bronson, D. Frendewey, W. Auerbach, D. Valenzuela, M. Karow, M. O. Hottiger, S. Hursting, J. C. Barrett, L. Guarente, R. Mulligan, B. Demple, G. D. Yancopoulos, F. W. Alt, Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 (2006). - PubMed
-
- Y. Kanfi, S. Naiman, G. Amir, V. Peshti, G. Zinman, L. Nahum, Z. Bar-Joseph, H. Y. Cohen, The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 (2012). - PubMed

