Supervised learning on impedance cytometry data for label-free biophysical distinction of pancreatic cancer cells versus their associated fibroblasts under gemcitabine treatment
- PMID: 37058962
- PMCID: PMC10134450
- DOI: 10.1016/j.bios.2023.115262
Supervised learning on impedance cytometry data for label-free biophysical distinction of pancreatic cancer cells versus their associated fibroblasts under gemcitabine treatment
Abstract
Chemotherapy failure in pancreatic cancer patients is widely attributed to cancer cell reprogramming towards drug resistance by cancer associated fibroblasts (CAFs), which are the abundant cell type in the tumor microenvironment. Association of drug resistance to specific cancer cell phenotypes within multicellular tumors can advance isolation protocols for enabling cell-type specific gene expression markers to identify drug resistance. This requires the distinction of drug resistant cancer cells versus CAFs, which is challenging since permeabilization of CAF cells during drug treatment can cause non-specific uptake of cancer cell-specific stains. Cellular biophysical metrics, on the other hand, can provide multiparametric information to assess the gradual alteration of target cancer cells towards drug resistance, but these phenotypes need to be distinguished versus CAFs. Using pancreatic cancer cells and CAFs from a metastatic patient-derived tumor that exhibits cancer cell drug resistance under CAF co-culture, the biophysical metrics from multifrequency single-cell impedance cytometry are utilized for distinction of the subpopulation of viable cancer cells versus CAFs, before and after gemcitabine treatment. This is accomplished through supervised machine learning after training the model using key impedance metrics for cancer cells and CAFs from transwell co-cultures, so that an optimized classifier model can recognize each cell type and predict their respective proportions in multicellular tumor samples, before and after gemcitabine treatment, as validated by their confusion matrix and flow cytometry assays. In this manner, an aggregate of the distinguishing biophysical metrics of viable cancer cells after gemcitabine treatment in co-cultures with CAFs can be used in longitudinal studies, to classify and isolate the drug resistant subpopulation for identifying markers.
Keywords: Chemotherapy; Impedance cytometry; Machine learning; Pancreatic cancer; Tumor microenvironment.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
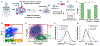
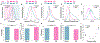





References
-
- AbuHammad and Zihlif 2013. Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line. Genomics 101(4):213–220. - PubMed
-
- Ahuja et al. 2019. Ahuja K; Rather GM; Lin Z; Sui J; Xie P; Le T; Bertino JR; Javanmard M, Toward point-of-care assessment of patient response: a portable tool for rapidly assessing cancer drug efficacy using multifrequency impedance cytometry and supervised machine learning. Microsystems & nanoengineering 5(1):1–11. - PMC - PubMed
-
- Caselli, et al. 2022. Caselli F; Reale R; De Ninno A; Spencer D; Morgan H; Bisegna P, Deciphering impedance cytometry signals with neural networks. Lab on a Chip 22(9):1714–1722. - PubMed
-
- Cheung, et al. 2010. Cheung KC; Di Berardino M; Schade-Kampmann G; Hebeisen M; Pierzchalski A; Bocsi J; Mittag A; Tárnok A, Microfluidic impedance-based flow cytometry. Cytometry part A 77(7):648–666. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

