A family-wide assessment of latent STAT transcription factor interactions reveals divergent dimer repertoires
- PMID: 37059181
- PMCID: PMC10200994
- DOI: 10.1016/j.jbc.2023.104703
A family-wide assessment of latent STAT transcription factor interactions reveals divergent dimer repertoires
Abstract
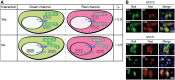
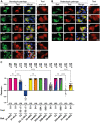
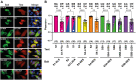
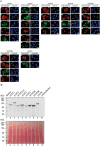
The conversion of signal transducer and activator of transcription (STAT) proteins from latent to active transcription factors is central to cytokine signaling. Triggered by their signal-induced tyrosine phosphorylation, it is the assembly of a range of cytokine-specific STAT homo- and heterodimers that marks a key step in the transition of hitherto latent proteins to transcription activators. In contrast, the constitutive self-assembly of latent STATs and how it relates to the functioning of activated STATs is understood less well. To provide a more complete picture, we developed a co-localization-based assay and tested all 28 possible combinations of the seven unphosphorylated STAT (U-STAT) proteins in living cells. We identified five U-STAT homodimers-STAT1, STAT3, STAT4, STAT5A, and STAT5B-and two heterodimers-STAT1:STAT2 and STAT5A:STAT5B-and performed semi-quantitative assessments of the forces and characterizations of binding interfaces that support them. One STAT protein-STAT6-was found to be monomeric. This comprehensive analysis of latent STAT self-assembly lays bare considerable structural and functional diversity in the ways that link STAT dimerization before and after activation.
Keywords: STAT1; STAT2; STAT3; STAT4; STAT5A; STAT5B; STAT6; dimerization; heterodimer; homodimer; interaction assay; nuclear export signal; nuclear import signal; transcription factor.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





Similar articles
-
JAK/STAT/SOCS-signaling pathway and colon and rectal cancer.Mol Carcinog. 2013 Feb;52(2):155-66. doi: 10.1002/mc.21841. Epub 2011 Nov 28. Mol Carcinog. 2013. PMID: 22121102 Free PMC article.
-
Interactions of the Nipah Virus P, V, and W Proteins across the STAT Family of Transcription Factors.mSphere. 2020 Dec 16;5(6):e00449-20. doi: 10.1128/mSphere.00449-20. mSphere. 2020. PMID: 33328346 Free PMC article.
-
Urokinase induces activation and formation of Stat4 and Stat1-Stat2 complexes in human vascular smooth muscle cells.J Biol Chem. 1999 Aug 20;274(34):24059-65. doi: 10.1074/jbc.274.34.24059. J Biol Chem. 1999. PMID: 10446176
-
Role of Jak kinases and STATs in cytokine signal transduction.Int J Hematol. 2001 Apr;73(3):271-7. doi: 10.1007/BF02981951. Int J Hematol. 2001. PMID: 11345192 Review.
-
The Dynamic Interface of Viruses with STATs.J Virol. 2020 Oct 27;94(22):e00856-20. doi: 10.1128/JVI.00856-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847860 Free PMC article. Review.
Cited by
-
The structural influence of the oncogenic driver mutation N642H in the STAT5B SH2 domain.Protein Sci. 2025 Jan;34(1):e70022. doi: 10.1002/pro.70022. Protein Sci. 2025. PMID: 39723827 Free PMC article.
-
eNAMPT/Ac-STAT3/DIRAS2 Axis Promotes Development and Cancer Stemness in Triple-Negative Breast Cancer by Enhancing Cytokine Crosstalk Between Tumor-Associated Macrophages and Cancer Cells.Int J Biol Sci. 2025 Feb 18;21(5):2027-2047. doi: 10.7150/ijbs.103723. eCollection 2025. Int J Biol Sci. 2025. PMID: 40083697 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

