Yolk-deprived Caenorhabditis elegans secure brood size at the expense of competitive fitness
- PMID: 37059473
- PMCID: PMC10105328
- DOI: 10.26508/lsa.202201675
Yolk-deprived Caenorhabditis elegans secure brood size at the expense of competitive fitness
Abstract
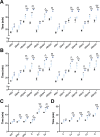
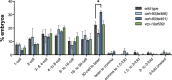
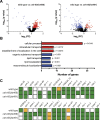
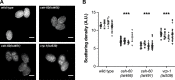
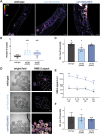
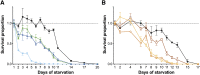
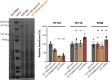
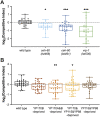
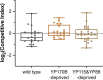
Oviparous animals support reproduction via the incorporation of yolk as a nutrient source into the eggs. In Caenorhabditis elegans, however, yolk proteins seem dispensable for fecundity, despite constituting the vast majority of the embryonic protein pool and acting as carriers for nutrient-rich lipids. Here, we used yolk protein-deprived C. elegans mutants to gain insight into the traits that may yet be influenced by yolk rationing. We show that massive yolk provisioning confers a temporal advantage during embryogenesis, while also increasing early juvenile body size and promoting competitive fitness. Opposite to species that reduce egg production under yolk deprivation, our results indicate that C. elegans relies on yolk as a fail-safe to secure offspring survival, rather than to maintain offspring numbers.
© 2023 Geens et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
