m6A-modification of cyclin D1 and c-myc IRESs in glioblastoma controls ITAF activity and resistance to mTOR inhibition
- PMID: 37061119
- PMCID: PMC10805108
- DOI: 10.1016/j.canlet.2023.216178
m6A-modification of cyclin D1 and c-myc IRESs in glioblastoma controls ITAF activity and resistance to mTOR inhibition
Abstract
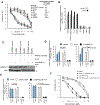
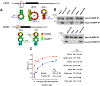
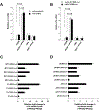
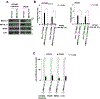
A major mechanism conferring resistance to mTOR inhibitors is activation of a salvage pathway stimulating internal ribosome entry site (IRES)-mediated mRNA translation, driving the synthesis of proteins promoting resistance of glioblastoma (GBM). Previously, we found this pathway is stimulated by the requisite IRES-trans-acting factor (ITAF) hnRNP A1, which itself is subject to phosphorylation and methylation events regulating cyclin D1 and c-myc IRES activity. Here we describe the requirement for m6A-modification of IRES RNAs for efficient translation and resistance to mTOR inhibition. DRACH-motifs within these IRES RNAs upon m6A modification resulted in enhanced IRES activity via increased hnRNP A1-binding following mTOR inhibitor exposure. Inhibitor exposure stimulated the expression of m6A-methylosome components resulting in increased activity in GBM. Silencing of METTL3-14 complexes reduced IRES activity upon inhibitor exposure and sensitized resistant GBM lines. YTHDF3 associates with m6A-modified cyclin D1 or c-myc IRESs, regulating IRES activity, and mTOR inhibitor sensitivity in vitro and in xenograft experiments. YTHDF3 interacted directly with hnRNP A1 and together stimulated hnRNP A1-dependent nucleic acid strand annealing activity. These data demonstrate that m6A-methylation of IRES RNAs regulate GBM responses to this class of inhibitors.
Keywords: Drug resistance; Glioblastoma; IRES; ITAF; N(6)-methyladenosine modification; mTOR inhibitors.
Published by Elsevier B.V.
Figures
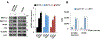

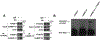
References
-
- Cloughesy TF, Cavenee WK, Mischel PS, Glioblastoma: from molecular pathology to targeted treatment, Annual review of pathology, 9 (2014) 1–25. - PubMed
-
- Omuro A, DeAngelis LM, Glioblastoma and other malignant gliomas: a clinical review, Jama, 310 (2013) 1842–1850. - PubMed
-
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, The somatic genomic landscape of glioblastoma, Cell, 155 (2013) 462–477. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

