SB Digestor: a tailored driver gene identification tool for dissecting heterogeneous Sleeping Beauty transposon-induced tumors
- PMID: 37063417
- PMCID: PMC10092771
- DOI: 10.7150/ijbs.81317
SB Digestor: a tailored driver gene identification tool for dissecting heterogeneous Sleeping Beauty transposon-induced tumors
Abstract
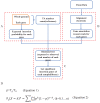
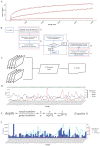
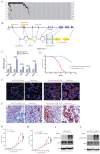
Sleeping Beauty (SB) insertional mutagenesis has been widely used for genome-wide functional screening in mouse models of human cancers, however, intertumor heterogeneity can be a major obstacle in identifying common insertion sites (CISs). Although previous algorithms have been successful in defining some CISs, they also miss CISs in certain situations. A major common characteristic of these previous methods is that they do not take tumor heterogeneity into account. However, intertumoral heterogeneity directly influences the sequence read number for different tumor samples and then affects CIS identification. To precisely detect and define cancer driver genes, we developed SB Digestor, a computational algorithm that overcomes biological heterogeneity to identify more potential driver genes. Specifically, we define the relationship between the sequenced read number and putative gene number to deduce the depth cutoff for each tumor, which can reduce tumor complexity and precisely reflect intertumoral heterogeneity. Using this new tool, we re-analyzed our previously published SB-based screening dataset and identified many additional potent drivers involved in Brca1-related tumorigenesis, including Arhgap42, Tcf12, and Fgfr2. SB Digestor not only greatly enhances our ability to identify and prioritize cancer drivers from SB tumors but also substantially deepens our understanding of the intrinsic genetic basis of cancer.
Keywords: Fgfr2; SB Digestor; Sleeping Beauty transposon; common insertion sites; intertumor heterogeneity.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
SB Driver Analysis: a Sleeping Beauty cancer driver analysis framework for identifying and prioritizing experimentally actionable oncogenes and tumor suppressors.Nucleic Acids Res. 2018 Sep 19;46(16):e94. doi: 10.1093/nar/gky450. Nucleic Acids Res. 2018. PMID: 29846651 Free PMC article.
-
Identification of Sleeping Beauty transposon insertions in solid tumors using linker-mediated PCR.J Vis Exp. 2013 Feb 1;(72):e50156. doi: 10.3791/50156. J Vis Exp. 2013. PMID: 23407503 Free PMC article.
-
Identification of cancer driver genes using Sleeping Beauty transposon mutagenesis.Cancer Sci. 2021 Jun;112(6):2089-2096. doi: 10.1111/cas.14901. Epub 2021 May 1. Cancer Sci. 2021. PMID: 33783919 Free PMC article. Review.
-
Sequencing methods and datasets to improve functional interpretation of sleeping beauty mutagenesis screens.BMC Genomics. 2014 Dec 19;15(1):1150. doi: 10.1186/1471-2164-15-1150. BMC Genomics. 2014. PMID: 25526783 Free PMC article.
-
Sleeping Beauty transposon insertional mutagenesis based mouse models for cancer gene discovery.Curr Opin Genet Dev. 2015 Feb;30:66-72. doi: 10.1016/j.gde.2015.04.007. Epub 2015 Jun 4. Curr Opin Genet Dev. 2015. PMID: 26051241 Free PMC article. Review.
References
-
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. - PubMed
-
- Weber J, Braun CJ, Saur D, Rad R. In vivo functional screening for systems-level integrative cancer genomics. Nat Rev Cancer. 2020;20:573–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

