Myeloid-specific blockade of Notch signaling ameliorates nonalcoholic fatty liver disease in mice
- PMID: 37063432
- PMCID: PMC10092768
- DOI: 10.7150/ijbs.80122
Myeloid-specific blockade of Notch signaling ameliorates nonalcoholic fatty liver disease in mice
Abstract
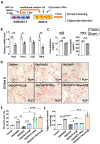
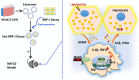
Rationale: Macrophages play a central role in the development and progression of nonalcoholic fatty liver disease (NAFLD). Studies have shown that Notch signaling mediated by transcription factor recombination signal binding protein for immunoglobulin kappa J region (RBP-J), is implicated in macrophage activation and plasticity. Naturally, we asked whether Notch signaling in macrophages plays a role in NAFLD, whether regulating Notch signaling in macrophages could serve as a therapeutic strategy to treat NAFLD. Methods: Immunofluorescence staining was used to detect the changes of macrophage Notch signaling in the livers of human patients with NAFLD and choline deficient amino acid-defined (CDAA) diet-fed mice. Lyz2-Cre RBP-Jflox or wild-type C57BL/6 male mice were fed with CDAA or high fat diet (HFD) to induce experimental steatohepatitis or steatosis, respectively. Liver histology examinations were performed using hematoxylin-eosin (H&E), Oil Red O staining, Sirius red staining and immunohistochemistry staining for F4/80, Col1α1 and αSMA. The expression of inflammatory factors, fibrosis or lipid metabolism associated genes were evaluated by quantitative reverse transcription (qRT)-PCR, Western blot or enzyme-linked immunosorbent assay (ELISA). The mRNA expression of liver samples was profiled by using RNA-seq. A hairpin-type decoy oligodeoxynucleotides (ODNs) for transcription factor RBP-J was loaded into bEnd.3-derived exosomes by electroporating. Mice with experimental NAFLD were treated with exosomes loading RBP-J decoy ODNs via tail vein injection. In vivo distribution of exosomes was analyzed by fluorescence labeling and imaging. Results: The results showed that Notch signaling was activated in hepatic macrophages in human with NAFLD or in CDAA-fed mice. Myeloid-specific RBP-J deficiency decreased the expression of inflammatory factors interleukin-1 beta (IL1β) and tumor necrosis factor alpha (TNFα), attenuated experimental steatohepatitis in mice. Furthermore, we found that Notch blockade attenuated lipid accumulation in hepatocytes by inhibiting the expression of IL1β and TNFα in macrophages in vitro. Meanwhile, we observed that tail vein-injected exosomes were mainly taken up by hepatic macrophages in mice with steatohepatitis. RBP-J decoy ODNs delivered by exosomes could efficiently inhibit Notch signaling in hepatic macrophages in vivo and ameliorate steatohepatitis or steatosis in CDAA or HFD mice, respectively. Conclusions: Combined, macrophage RBP-J promotes the progression of NAFLD at least partially through regulating the expression of pro-inflammatory cytokines IL1β and TNFα. Infusion of exosomes loaded with RBP-J decoy ODNs might be a promising therapy to treat NAFLD.
Keywords: Notch signaling; RBP-J; exosomes; macrophage; nonalcoholic fatty liver disease; transcription factor decoy.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. - PubMed
-
- Diehl AM, Day C. Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. N Engl J Med. 2017;377:2063–2072. - PubMed
-
- Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–2224. - PubMed
-
- Heymann F, Tacke F. Immunology in the liver-from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13:88–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

