Identifying TNF and IL6 as potential hub genes and targeted drugs associated with scleritis: A bio-informative report
- PMID: 37063831
- PMCID: PMC10102337
- DOI: 10.3389/fimmu.2023.1098140
Identifying TNF and IL6 as potential hub genes and targeted drugs associated with scleritis: A bio-informative report
Abstract
Background: Scleritis is a serious inflammatory eye disease that can lead to blindness. The etiology and pathogenesis of scleritis remain unclear, and increasing evidence indicates that some specific genes and proteins are involved. This study aimed to identify pivotal genes and drug targets for scleritis, thus providing new directions for the treatment of this disease.
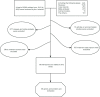
Methods: We screened candidate genes and proteins associated with scleritis by text-mining the PubMed database using Python, and assessed their functions by using the DAVID database. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used to identify the functional enrichment of these genes and proteins. Then, the hub genes were identified with CytoHubba and assessed by protein-protein interaction (PPI) network analysis. And the serum from patients with active scleritis and healthy subjects were used for the validation of hub genes. Finally, the DGIdb database was used to predict targeted drugs for the hub genes for treating scleritis.
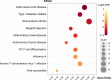
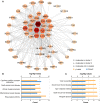
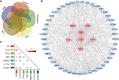
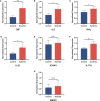
Results: A total of 56 genes and proteins were found to be linked to scleritis, and 65 significantly altered pathways were identified in the KEGG analysis (FDR < 0.05). Most of the top five pathways involved the categories "Rheumatoid arthritis," "Inflammatory bowel disease", "Type I diabetes mellitus," and "Graft-versus-host disease". TNF and IL6 were considered to be the top 2 hub genes through CytoHubba. Based on our serum samples, hub genes are expressed at high levels in active scleritis. Five scleritis-targeting drugs were found among 88 identified drugs.
Conclusions: This study provides key genes and drug targets related to scleritis through bioinformatics analysis. TNF and IL6 are considered key mediators and possible drug targets of scleritis. Five drug candidates may play an important role in the diagnosis and treatment of scleritis in the future, which is worthy of the further experimental and clinical study.
Keywords: IL6; PPI network; TNF; drug targets; hub genes; scleritis.
Copyright © 2023 Yan, Liu, Zhang and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

